bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2004_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
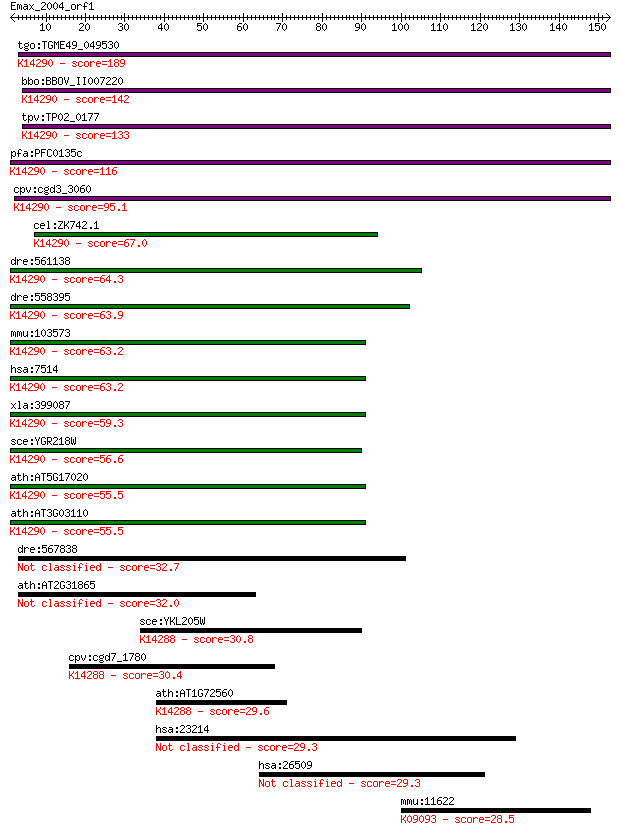
tgo:TGME49_049530 exportin, putative ; K14290 exportin-1 189 4e-48
bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1 142 4e-34
tpv:TP02_0177 importin beta-related nuclear transport factor; ... 133 3e-31
pfa:PFC0135c exportin 1, putative; K14290 exportin-1 116 3e-26
cpv:cgd3_3060 exportin 1 ; K14290 exportin-1 95.1 8e-20
cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family m... 67.0 2e-11
dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b; K1... 64.3 1e-10
dre:558395 xpo1a, im:7151655; exportin 1 (CRM1 homolog, yeast)... 63.9 2e-10
mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolo... 63.2 3e-10
hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homo... 63.2 3e-10
xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog); ... 59.3 4e-09
sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1 56.6 3e-08
ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transpor... 55.5 6e-08
ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14... 55.5 6e-08
dre:567838 MGC172359, heatr6; zgc:172359 32.7 0.42
ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family p... 32.0 0.71
sce:YKL205W LOS1; Los1p; K14288 exportin-T 30.8 1.9
cpv:cgd7_1780 tRNA exportin type nuclear export protein ; K142... 30.4 2.2
ath:AT1G72560 PSD; PSD (PAUSED); nucleobase, nucleoside, nucle... 29.6 4.2
hsa:23214 XPO6, EXP6, FLJ22519, KIAA0370, RANBP20; exportin 6 29.3
hsa:26509 MYOF, FER1L3, FLJ36571, FLJ90777; myoferlin 29.3 5.5
mmu:11622 Ahr, Ah, Ahh, Ahre, In, bHLHe76; aryl-hydrocarbon re... 28.5 8.5
> tgo:TGME49_049530 exportin, putative ; K14290 exportin-1
Length=1125
Score = 189 bits (479), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 87/150 (58%), Positives = 118/150 (78%), Gaps = 1/150 (0%)
Query 3 ELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELLPL 62
+LT QF+E+++VC+FVLKSF+ + +K +L+QQTLKCLAHFLKWIPLGFVFET+L+
Sbjct 194 QLTAQFQEVFDVCMFVLKSFVVNA-AAMKESLVQQTLKCLAHFLKWIPLGFVFETDLIET 252
Query 63 LLQYFYEDVSFRIDCLRCLTEVSSLQLNINELNLFSQQMTLLWAQLSDKAAHTPKQSLQY 122
LLQ+F+E V FR DCLRC+TE++SLQL+ E +F +++ +LW +L K P Q+L++
Sbjct 253 LLQHFWEPVQFRADCLRCVTEIASLQLSKEETQVFRERLAVLWLELVGKVLALPPQTLRF 312
Query 123 NDPTSVPPQVRLFWETTFCQLELCLTAFLR 152
D VPPQ+RLFWET +CQL LCLTAFL+
Sbjct 313 EDAGQVPPQMRLFWETIYCQLSLCLTAFLK 342
> bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1
Length=1186
Score = 142 bits (358), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 61/157 (38%), Positives = 104/157 (66%), Gaps = 9/157 (5%)
Query 4 LTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFE------- 56
++ FK+++ +CIFV+ + I++P ++++L++QTL CLAHFLKWIP+G++FE
Sbjct 193 MSADFKDIFELCIFVMHNSITNPES-VRVSLVKQTLTCLAHFLKWIPVGYIFEQYFYGGV 251
Query 57 -TELLPLLLQYFYEDVSFRIDCLRCLTEVSSLQLNINELNLFSQQMTLLWAQLSDKAAHT 115
L+ LLL +F++ +++R++C +CLTE++ L L+ E+ F ++ +W +L K +
Sbjct 252 NVVLIDLLLDHFWDSMTYRVECTKCLTEIAGLSLSSQEMQAFGMRVASMWPKLVAKVSSL 311
Query 116 PKQSLQYNDPTSVPPQVRLFWETTFCQLELCLTAFLR 152
P+ S Y+D V P RLFWET +CQ +C T FL+
Sbjct 312 PENSTHYDDTNHVAPCNRLFWETFYCQFSICCTNFLK 348
> tpv:TP02_0177 importin beta-related nuclear transport factor;
K14290 exportin-1
Length=1067
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 71/157 (45%), Positives = 106/157 (67%), Gaps = 10/157 (6%)
Query 4 LTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFET------ 57
+T +F+E++ +CIFVL SFIS+P+ + TL++QTL CL+HFLKWIP G++FE+
Sbjct 197 MTSEFREIFELCIFVLNSFISNPNM-VNNTLVKQTLVCLSHFLKWIPYGYIFESYPHGEG 255
Query 58 --ELLPLLLQYFYEDVSFRIDCLRCLTEVSSLQLNINELNLFSQQMTLLWAQLSDKAAHT 115
LL LLL +F++ +++R++C +CL EV+SL L NEL FS ++ LW ++ K +
Sbjct 256 SVVLLDLLLDHFWDPMTYRVECTKCLNEVASLTLTNNELQTFSHRIMSLWPKVVQKVSTL 315
Query 116 PKQSLQYNDPTSVPPQVRLFWETTFCQLELCLTAFLR 152
P +S QY D +PP + LFWET + QL L LT L+
Sbjct 316 PPESFQY-DNAKIPPSMLLFWETFYTQLTLFLTNLLK 351
> pfa:PFC0135c exportin 1, putative; K14290 exportin-1
Length=1254
Score = 116 bits (291), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 52/160 (32%), Positives = 100/160 (62%), Gaps = 13/160 (8%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFET--- 57
R+E QF+E+YN+C+++L++ + + +LI+QTL CL++F KWIPL ++F+
Sbjct 194 RNEYASQFQEVYNLCLYILEANVYNKRST-NTSLIKQTLHCLSNFFKWIPLTYIFDKYKF 252
Query 58 -----ELLPLLLQYFYEDVSFRIDCLRCLTEVSSLQLNINELNLFSQQMTLLWAQLSDKA 112
+++ LL +F++D+S++I+C++C+ E+ L+++ + F LW +L K
Sbjct 253 NDNNIQIIDLLFDHFWDDISYKIECVKCIQEIVMLKIDEKNILYFDNVFINLWTKLVSKI 312
Query 113 AHTPKQSLQYNDPTSVPPQVRLFWETTFCQLELCLTAFLR 152
P N+ ++PP++++FWE F QL +C+T+FL+
Sbjct 313 KLLPNA----NEMKNIPPELKIFWEQYFLQLSICITSFLK 348
> cpv:cgd3_3060 exportin 1 ; K14290 exportin-1
Length=1266
Score = 95.1 bits (235), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 52/178 (29%), Positives = 94/178 (52%), Gaps = 28/178 (15%)
Query 2 DELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELLP 61
D L +QF ++ ++ +FVL S++ +P IK+ L+ +L+CL H+LKWIPL ++ E +L P
Sbjct 189 DILNQQFPQILSLILFVLTSYLENPQN-IKVNLVVSSLQCLCHYLKWIPLNYILECDLRP 247
Query 62 --------------------LLLQYFYEDVSFRIDCLRCLTEVSSLQLNINELNL----- 96
LL +F+ + SFR++ ++CLTE+S L+ + N +
Sbjct 248 QLPHSIASNGNNNIIYNLLQFLLDHFWGNPSFRLESIKCLTEISPLKFDENTKDSNGGLN 307
Query 97 --FSQQMTLLWAQLSDKAAHTPKQSLQYNDPTSVPPQVRLFWETTFCQLELCLTAFLR 152
QM +W + ++ P + QY+ +V RL++E F + L L++F++
Sbjct 308 KQIEDQMVQIWLSIVNRIKEVPNEYAQYDSMPNVSTSTRLYYERYFNYIALLLSSFIK 365
> cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family
member (xpo-1); K14290 exportin-1
Length=1080
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 55/87 (63%), Gaps = 7/87 (8%)
Query 7 QFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELLPLLLQY 66
QF+E++ +C+ +L+ S+ +++Q TLK L FL WIP+G+VFET + LL +
Sbjct 200 QFQEVFTLCVSILEKCPSN-------SMVQATLKTLQRFLTWIPVGYVFETNITELLSEN 252
Query 67 FYEDVSFRIDCLRCLTEVSSLQLNINE 93
F +R+ L+CLTE+S +Q+ N+
Sbjct 253 FLSLEVYRVIALQCLTEISQIQVETND 279
> dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b;
K14290 exportin-1
Length=1071
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/107 (33%), Positives = 63/107 (58%), Gaps = 10/107 (9%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+D + +F +++ +C FV+++ ++P L+ TL+ L FL WIPLG++FET+L+
Sbjct 195 KDSMCNEFSQIFQLCQFVMENSQNAP-------LVHATLETLLRFLNWIPLGYIFETKLI 247
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLNINE---LNLFSQQMTLL 104
L+ F FR L+CLTE++ + ++ E +NLF+ M L
Sbjct 248 STLVYKFLNVPMFRNVTLKCLTEIAGVSVSQYEEQFVNLFTLTMMQL 294
> dre:558395 xpo1a, im:7151655; exportin 1 (CRM1 homolog, yeast)
a; K14290 exportin-1
Length=687
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 62/104 (59%), Gaps = 10/104 (9%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+D + +F +++ +C FV+++ ++P L+ TL+ L FL WIPLG++FET+L+
Sbjct 195 KDSMCNEFSQIFQLCQFVMENSQNAP-------LVHATLETLLRFLNWIPLGYIFETKLI 247
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLNINE---LNLFSQQM 101
L+ F FR L+CLTE++ + ++ E +NLF+ M
Sbjct 248 STLVYKFLNVPMFRNVTLKCLTEIAGVSVSQYEEQFVNLFTLTM 291
> mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolog
(yeast); K14290 exportin-1
Length=1071
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 55/90 (61%), Gaps = 7/90 (7%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+D + +F +++ +C FV+++ ++P L+ TL+ L FL WIPLG++FET+L+
Sbjct 195 KDSMCNEFSQIFQLCQFVMENSQNAP-------LVHATLETLLRFLNWIPLGYIFETKLI 247
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLN 90
L+ F FR L+CLTE++ + ++
Sbjct 248 STLIYKFLNVPMFRNVSLKCLTEIAGVSVS 277
> hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homolog,
yeast); K14290 exportin-1
Length=1071
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 55/90 (61%), Gaps = 7/90 (7%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+D + +F +++ +C FV+++ ++P L+ TL+ L FL WIPLG++FET+L+
Sbjct 195 KDSMCNEFSQIFQLCQFVMENSQNAP-------LVHATLETLLRFLNWIPLGYIFETKLI 247
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLN 90
L+ F FR L+CLTE++ + ++
Sbjct 248 STLIYKFLNVPMFRNVSLKCLTEIAGVSVS 277
> xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog);
K14290 exportin-1
Length=1071
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 54/90 (60%), Gaps = 7/90 (7%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+D + +F +++ +C FV+++ ++ L+ TL+ L FL WIPLG++FET+L+
Sbjct 195 KDSMCNEFSQIFQLCQFVMENSQNA-------QLVHATLETLLRFLNWIPLGYIFETKLI 247
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLN 90
L+ F FR L+CLTE++ + ++
Sbjct 248 STLVYKFLNVPMFRNVSLKCLTEIAGVSVS 277
> sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1
Length=1084
Score = 56.6 bits (135), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 53/89 (59%), Gaps = 7/89 (7%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
++ ++K+F++++ +C VL+ SS +LI TL+ L +L WIP +++ET +L
Sbjct 183 KNSMSKEFEQIFKLCFQVLEQGSSS-------SLIVATLESLLRYLHWIPYRYIYETNIL 235
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQL 89
LL F R L+CLTEVS+L++
Sbjct 236 ELLSTKFMTSPDTRAITLKCLTEVSNLKI 264
> ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transporter/
receptor; K14290 exportin-1
Length=1075
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 53/90 (58%), Gaps = 8/90 (8%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+ L +FK ++ +C++VL + + LI+ TL L +L WIPLG++FE+ LL
Sbjct 186 KQSLNSEFKLIHELCLYVLSA-------SQRQDLIRATLSALHAYLSWIPLGYIFESTLL 238
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLN 90
LL+ F+ ++R ++CLTEV++L
Sbjct 239 ETLLK-FFPVPAYRNLTIQCLTEVAALNFG 267
> ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14290
exportin-1
Length=1076
Score = 55.5 bits (132), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 54/90 (60%), Gaps = 8/90 (8%)
Query 1 RDELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELL 60
+ L +F+ ++ +C++VL + + LI+ TL L +L WIPLG++FE+ LL
Sbjct 186 KQSLNSEFQLIHELCLYVLSA-------SQRQELIRATLSALHAYLSWIPLGYIFESPLL 238
Query 61 PLLLQYFYEDVSFRIDCLRCLTEVSSLQLN 90
+LL+ F+ ++R L+CL+EV+SL
Sbjct 239 EILLK-FFPVPAYRNLTLQCLSEVASLNFG 267
> dre:567838 MGC172359, heatr6; zgc:172359
Length=1201
Score = 32.7 bits (73), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 45/110 (40%), Gaps = 12/110 (10%)
Query 3 ELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQ--------QTLKCLAHFLKWIPLGFV 54
E T ++ + L + I H G+ + LI Q LKCLAH + +P +
Sbjct 504 EDTGAPRQAFTPFSATLAASIRELHRGLLLALIAESSCQTLTQVLKCLAHLVSNVPYNRL 563
Query 55 FETELLPLLLQ----YFYEDVSFRIDCLRCLTEVSSLQLNINELNLFSQQ 100
L PL Q + DV+ R+ L + S Q + E+ L QQ
Sbjct 564 RPGLLSPLWKQIRPYVRHRDVNVRVSSLTLFGALVSTQAPLPEVQLLLQQ 613
> ath:AT2G31865 poly (ADP-ribose) glycohydrolase (PARG) family
protein
Length=522
Score = 32.0 bits (71), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 32/65 (49%), Gaps = 9/65 (13%)
Query 3 ELTKQFKELYNVCIFVLKSFISSPHGGIKITLIQQTLKCLAHFL----KWIPLGFV-FET 57
E+ + K L + L SF H T + +KCL H+ +W+P GFV FE
Sbjct 162 EVDRSLKNLQGINFSGLFSFPYMRH----CTKQENKIKCLIHYFGRICRWMPTGFVSFER 217
Query 58 ELLPL 62
++LPL
Sbjct 218 KILPL 222
> sce:YKL205W LOS1; Los1p; K14288 exportin-T
Length=1100
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 28/57 (49%), Gaps = 1/57 (1%)
Query 34 LIQQTLKCLAHFLKWIPLGFVFETELLPLLLQYFYEDVS-FRIDCLRCLTEVSSLQL 89
LI TL C+ F+ WI + + + L L Y + ++ +I C C+ + S ++
Sbjct 226 LINSTLDCIGSFISWIDINLIIDANNYYLQLIYKFLNLKETKISCYNCILAIISKKM 282
> cpv:cgd7_1780 tRNA exportin type nuclear export protein ; K14288
exportin-T
Length=1303
Score = 30.4 bits (67), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 13/52 (25%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 16 IFVLKSFISSPHGGIKITLIQQTLKCLAHFLKWIPLGFVFETELLPLLLQYF 67
IFV+ S +I L+ ++K + F+ WI + + E+L ++L +
Sbjct 212 IFVVSKHKSQSLTKDRIQLLSSSMKMMESFIDWIDISYAVNNEVLSIILSFL 263
> ath:AT1G72560 PSD; PSD (PAUSED); nucleobase, nucleoside, nucleotide
and nucleic acid transmembrane transporter/ tRNA binding;
K14288 exportin-T
Length=988
Score = 29.6 bits (65), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 16/33 (48%), Gaps = 0/33 (0%)
Query 38 TLKCLAHFLKWIPLGFVFETELLPLLLQYFYED 70
L C+ F+ WI +G V +PLL + D
Sbjct 213 VLDCMRRFVSWIDIGLVANDAFVPLLFELILSD 245
> hsa:23214 XPO6, EXP6, FLJ22519, KIAA0370, RANBP20; exportin
6
Length=1125
Score = 29.3 bits (64), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 30/120 (25%), Positives = 44/120 (36%), Gaps = 29/120 (24%)
Query 38 TLKCLAHFLKWIPLGFVFETELLPLLLQY--FYEDVSFR--------------------- 74
L+CLAH WIPL LL + + F D+ R
Sbjct 249 ALECLAHLFSWIPLSASITPSLLTTIFHFARFGCDIRARKMASVNGSSQNCVSGQERGRL 308
Query 75 -IDCLRCLTEVSS---LQLNINE--LNLFSQQMTLLWAQLSDKAAHTPKQSLQYNDPTSV 128
+ + C+ E+ S + + E L +F Q LL D AHT K L+ D + +
Sbjct 309 GVLAMSCINELMSKNCVPMEFEEYLLRMFQQTFYLLQKITKDNNAHTVKSRLEELDESYI 368
> hsa:26509 MYOF, FER1L3, FLJ36571, FLJ90777; myoferlin
Length=2061
Score = 29.3 bits (64), Expect = 5.5, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 32/64 (50%), Gaps = 7/64 (10%)
Query 64 LQYFYEDVSFRIDCLRCLTEVSS-LQLNINEL------NLFSQQMTLLWAQLSDKAAHTP 116
L ++ED+S R+D + L ++ LQ NI L + + Q+ LW +L D+
Sbjct 643 LTSYWEDISHRLDAVNTLLAMAERLQTNIEALKSGIQGKIPANQLAELWLKLIDEVIEDT 702
Query 117 KQSL 120
+ +L
Sbjct 703 RYTL 706
> mmu:11622 Ahr, Ah, Ahh, Ahre, In, bHLHe76; aryl-hydrocarbon
receptor; K09093 aryl hydrocarbon receptor
Length=805
Score = 28.5 bits (62), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 25/54 (46%), Gaps = 7/54 (12%)
Query 100 QMTLLWAQLSDKA-----AHTPKQSLQYNDPTSVPPQVRLFWETTF-CQLELCL 147
Q L WA D A AH P Q+ Y P +PP+ F E F C+L CL
Sbjct 169 QRQLHWALNPDSAQGVDEAHGPPQAAVYYTPDQLPPENASFMERCFRCRLR-CL 221
Lambda K H
0.327 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40