bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
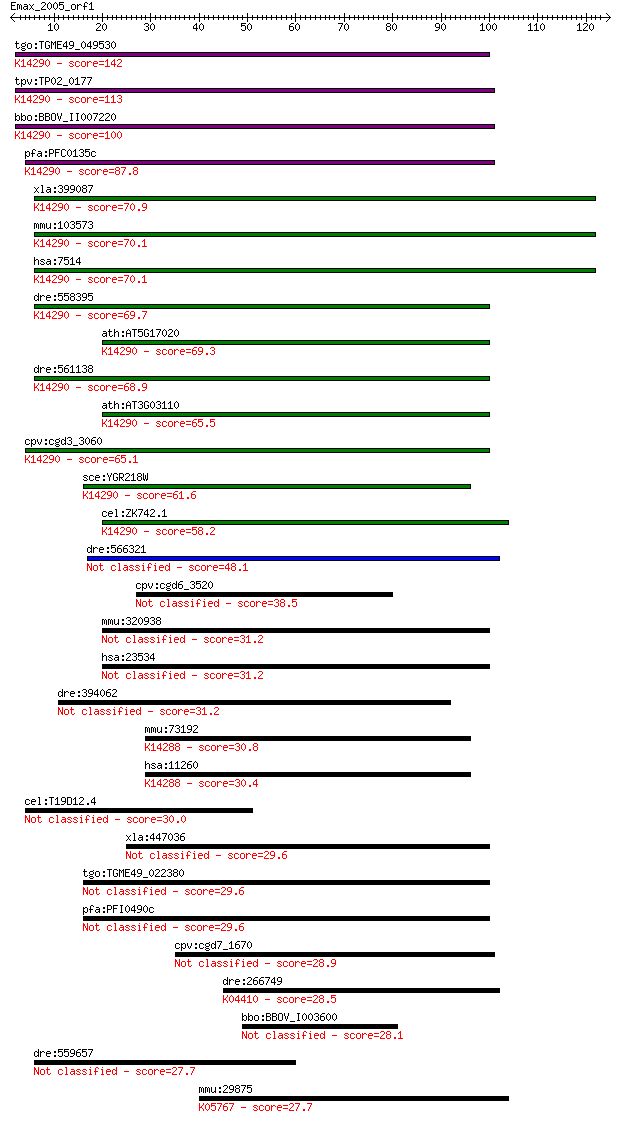
Query= Emax_2005_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_049530 exportin, putative ; K14290 exportin-1 142 3e-34
tpv:TP02_0177 importin beta-related nuclear transport factor; ... 113 2e-25
bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1 100 1e-21
pfa:PFC0135c exportin 1, putative; K14290 exportin-1 87.8 8e-18
xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog); ... 70.9 9e-13
mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolo... 70.1 2e-12
hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homo... 70.1 2e-12
dre:558395 xpo1a, im:7151655; exportin 1 (CRM1 homolog, yeast)... 69.7 2e-12
ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transpor... 69.3 3e-12
dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b; K1... 68.9 4e-12
ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14... 65.5 4e-11
cpv:cgd3_3060 exportin 1 ; K14290 exportin-1 65.1 5e-11
sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1 61.6 5e-10
cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family m... 58.2 7e-09
dre:566321 importin-13-like 48.1 6e-06
cpv:cgd6_3520 hypothetical protein 38.5 0.006
mmu:320938 Tnpo3, 5730544L10Rik, C430013M08Rik, C81142, D6Ertd... 31.2 0.77
hsa:23534 TNPO3, IPO12, MTR10A, TRN-SR, TRN-SR2, TRNSR; transp... 31.2 0.78
dre:394062 tnpo3, MGC55409, zgc:55409; transportin 3 31.2
mmu:73192 Xpot, 1110004L07Rik, 3110065H13Rik, AI452076, C79645... 30.8 1.1
hsa:11260 XPOT, XPO3; exportin, tRNA (nuclear export receptor ... 30.4 1.3
cel:T19D12.4 hypothetical protein 30.0 2.0
xla:447036 tnpo3, MGC82948; transportin 3 29.6
tgo:TGME49_022380 exportin 7, putative 29.6 2.4
pfa:PFI0490c ran-binding protein, putative 29.6 2.8
cpv:cgd7_1670 nuclear pore protein RBP16/17 (RAN binding prote... 28.9 4.2
dre:266749 pak2a, cb422, kinase, pak2, si:dz198m22.1, wu:fb71h... 28.5 5.3
bbo:BBOV_I003600 19.m02390; hypothetical protein 28.1 7.3
dre:559657 zgc:101095; si:dkeyp-11g8.6 27.7 9.0
mmu:29875 Iqgap1, AA682088, D7Ertd237e, D7Ertd257e, mKIAA0051;... 27.7 9.6
> tgo:TGME49_049530 exportin, putative ; K14290 exportin-1
Length=1125
Score = 142 bits (358), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 67/98 (68%), Positives = 80/98 (81%), Gaps = 0/98 (0%)
Query 2 LTDPRALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAF 61
L+DPRALLD ++PF++A V LLD+VVAAMF + D+ RDVAH+VLGEFKNMP AW++VA
Sbjct 3 LSDPRALLDASIPFDDAKVALLDQVVAAMFGTTDNHSRDVAHKVLGEFKNMPEAWSYVAV 62
Query 62 ILNVSKDPNTKFFALQILEATITNRWNVLPETERNSTK 99
ILN S+D NTKF ALQILE TI RWNVLP+ ERN K
Sbjct 63 ILNKSQDANTKFVALQILENTIQTRWNVLPDAERNGIK 100
> tpv:TP02_0177 importin beta-related nuclear transport factor;
K14290 exportin-1
Length=1067
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 52/99 (52%), Positives = 67/99 (67%), Gaps = 0/99 (0%)
Query 2 LTDPRALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAF 61
LTDP LLD + PF+EA V LLD V+ +MF R+ AHR+L +FK +P++W HVA
Sbjct 5 LTDPLVLLDTSRPFDEAMVPLLDSVIISMFDGTSVENRETAHRILEQFKKLPDSWKHVAL 64
Query 62 ILNVSKDPNTKFFALQILEATITNRWNVLPETERNSTKS 100
IL S + NTKF+ALQ+LE I RWN+LP+TER K
Sbjct 65 ILAKSSNSNTKFYALQVLEICIETRWNILPDTERAGIKQ 103
> bbo:BBOV_II007220 18.m06600; exportin 1; K14290 exportin-1
Length=1186
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 47/99 (47%), Positives = 65/99 (65%), Gaps = 0/99 (0%)
Query 2 LTDPRALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAF 61
+ DP LLD + F+E V LLD V+ AMF S R+ AH++L +F+ +P++W HVA
Sbjct 1 MADPSILLDTSRVFDENMVALLDTVIDAMFDSGSGHNREAAHKILEQFRTLPDSWKHVAV 60
Query 62 ILNVSKDPNTKFFALQILEATITNRWNVLPETERNSTKS 100
IL+ SK+ NTKFFALQ+L+ I RWNVL +R +S
Sbjct 61 ILSCSKNTNTKFFALQVLQMCIQTRWNVLAIEDRLGIRS 99
> pfa:PFC0135c exportin 1, putative; K14290 exportin-1
Length=1254
Score = 87.8 bits (216), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 42/97 (43%), Positives = 59/97 (60%), Gaps = 0/97 (0%)
Query 4 DPRALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFIL 63
+P +LLD N F+ + LLD VV A+ + D RD A +L +FK + +W V+ IL
Sbjct 7 NPLSLLDKNQAFDAEKLKLLDNVVEALLDTKDKNRRDFAQNLLNQFKMLDTSWRSVSIIL 66
Query 64 NVSKDPNTKFFALQILEATITNRWNVLPETERNSTKS 100
S++ NTKF+ LQILE I NRWN+LP E+ K+
Sbjct 67 EHSENVNTKFYGLQILEECINNRWNILPSEEKEGMKN 103
> xla:399087 xpo1, crm1, exportin-1; exportin 1 (CRM1 homolog);
K14290 exportin-1
Length=1071
Score = 70.9 bits (172), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 65/123 (52%), Gaps = 11/123 (8%)
Query 6 RALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNV 65
R LLD + + +NLLD VV ++ + +R +A VL K P+AWT V IL
Sbjct 14 RQLLDFSQKLD---INLLDNVVNCLYHGEGAQQR-MAQEVLTHLKEHPDAWTRVDTILEF 69
Query 66 SKDPNTKFFALQILEATITNRWNVLPETERNSTKS-------RSIKNKTCGYKMFIYMQK 118
S++ NTK++ LQILE I RW +LP + + K ++ + TC K +Y+ K
Sbjct 70 SQNMNTKYYGLQILENVIKTRWKILPRNQCDGIKKYVVGLIIKTSSDATCVEKEKVYIGK 129
Query 119 GNI 121
N+
Sbjct 130 LNM 132
> mmu:103573 Xpo1, AA420417, Crm1, Exp1; exportin 1, CRM1 homolog
(yeast); K14290 exportin-1
Length=1071
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 64/123 (52%), Gaps = 11/123 (8%)
Query 6 RALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNV 65
R LLD + + +NLLD VV ++ + +R +A VL K P+AWT V IL
Sbjct 14 RQLLDFSQKLD---INLLDNVVNCLYHGEGAQQR-MAQEVLTHLKEHPDAWTRVDTILEF 69
Query 66 SKDPNTKFFALQILEATITNRWNVLPETERNSTKS-------RSIKNKTCGYKMFIYMQK 118
S++ NTK++ LQILE I RW +LP + K ++ + TC K +Y+ K
Sbjct 70 SQNMNTKYYGLQILENVIKTRWKILPRNQCEGIKKYVVGLIIKTSSDPTCVEKEKVYIGK 129
Query 119 GNI 121
N+
Sbjct 130 LNM 132
> hsa:7514 XPO1, CRM1, DKFZp686B1823, emb; exportin 1 (CRM1 homolog,
yeast); K14290 exportin-1
Length=1071
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 64/123 (52%), Gaps = 11/123 (8%)
Query 6 RALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNV 65
R LLD + + +NLLD VV ++ + +R +A VL K P+AWT V IL
Sbjct 14 RQLLDFSQKLD---INLLDNVVNCLYHGEGAQQR-MAQEVLTHLKEHPDAWTRVDTILEF 69
Query 66 SKDPNTKFFALQILEATITNRWNVLPETERNSTKS-------RSIKNKTCGYKMFIYMQK 118
S++ NTK++ LQILE I RW +LP + K ++ + TC K +Y+ K
Sbjct 70 SQNMNTKYYGLQILENVIKTRWKILPRNQCEGIKKYVVGLIIKTSSDPTCVEKEKVYIGK 129
Query 119 GNI 121
N+
Sbjct 130 LNM 132
> dre:558395 xpo1a, im:7151655; exportin 1 (CRM1 homolog, yeast)
a; K14290 exportin-1
Length=687
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 54/94 (57%), Gaps = 4/94 (4%)
Query 6 RALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNV 65
R LLD + + +NLLD VV +M+ S +R +A VL K+ P+AWT V IL
Sbjct 14 RQLLDFSQKLD---INLLDNVVNSMYYDVGSQQR-LAQEVLTNLKDHPDAWTRVDTILEF 69
Query 66 SKDPNTKFFALQILEATITNRWNVLPETERNSTK 99
S++ TK++ALQILE I RW +LP + K
Sbjct 70 SQNMKTKYYALQILETVIKTRWKILPRNQCEGIK 103
> ath:AT5G17020 XPO1A; XPO1A; protein binding / protein transporter/
receptor; K14290 exportin-1
Length=1075
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 46/80 (57%), Gaps = 0/80 (0%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
V +LD VAA F++ ER A ++L + + P+ W V IL + +TKFFALQ+L
Sbjct 15 VGVLDATVAAFFVTGSKEERAAADQILRDLQANPDMWLQVVHILQNTNSLDTKFFALQVL 74
Query 80 EATITNRWNVLPETERNSTK 99
E I RWN LP +R+ K
Sbjct 75 EGVIKYRWNALPVEQRDGMK 94
> dre:561138 xpo1b, xpo1; exportin 1 (CRM1 homolog, yeast) b;
K14290 exportin-1
Length=1071
Score = 68.9 bits (167), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 39/94 (41%), Positives = 52/94 (55%), Gaps = 4/94 (4%)
Query 6 RALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNV 65
R LLD N + +NLLD VV ++ +R +A VL K P+AWT V IL
Sbjct 14 RQLLDFNQKLD---INLLDNVVNCLYHGVGPQQR-MAQEVLTHLKEHPDAWTRVDTILEF 69
Query 66 SKDPNTKFFALQILEATITNRWNVLPETERNSTK 99
S++ NTK++ALQILE I RW +LP + K
Sbjct 70 SQNMNTKYYALQILETVIKTRWKILPRNQCEGIK 103
> ath:AT3G03110 XPO1B; XPO1B; binding / protein transporter; K14290
exportin-1
Length=1076
Score = 65.5 bits (158), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 43/80 (53%), Gaps = 0/80 (0%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
V LLD V A + + ER A +L + K P+ W V IL + +TKFFALQ+L
Sbjct 15 VVLLDATVEAFYSTGSKEERASADNILRDLKANPDTWLQVVHILQNTSSTHTKFFALQVL 74
Query 80 EATITNRWNVLPETERNSTK 99
E I RWN LP +R+ K
Sbjct 75 EGVIKYRWNALPVEQRDGMK 94
> cpv:cgd3_3060 exportin 1 ; K14290 exportin-1
Length=1266
Score = 65.1 bits (157), Expect = 5e-11, Method: Composition-based stats.
Identities = 36/96 (37%), Positives = 53/96 (55%), Gaps = 1/96 (1%)
Query 4 DPRALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFIL 63
D LLD + P++ V +LD +V M+ +R +A ++L E K ++W V IL
Sbjct 2 DISLLLDLSQPYDLQKVEMLDELVGVMYGLRPG-DRIIADKILSELKQKTDSWRIVGNIL 60
Query 64 NVSKDPNTKFFALQILEATITNRWNVLPETERNSTK 99
+S D NTKFFAL ILE I +W +LP ++ K
Sbjct 61 QLSSDYNTKFFALSILEKCIQFQWKILPFDQKTGIK 96
> sce:YGR218W CRM1, KAP124, XPO1; Crm1p; K14290 exportin-1
Length=1084
Score = 61.6 bits (148), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 46/80 (57%), Gaps = 1/80 (1%)
Query 16 NEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFA 75
N+ + LLD+VV+ F V++ A +L +F++ P+AW IL S +P +KF A
Sbjct 9 NDLDIALLDQVVST-FYQGSGVQQKQAQEILTKFQDNPDAWQKADQILQFSTNPQSKFIA 67
Query 76 LQILEATITNRWNVLPETER 95
L IL+ IT +W +LP R
Sbjct 68 LSILDKLITRKWKLLPNDHR 87
> cel:ZK742.1 xpo-1; eXPOrtin (nuclear export receptor) family
member (xpo-1); K14290 exportin-1
Length=1080
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 33/84 (39%), Positives = 48/84 (57%), Gaps = 1/84 (1%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
V LLD+VV M + E+ A+++L K ++WT V IL S+ +K+FALQIL
Sbjct 24 VTLLDQVVEIMNRMSGK-EQAEANQILMSLKEERDSWTKVDAILQYSQLNESKYFALQIL 82
Query 80 EATITNRWNVLPETERNSTKSRSI 103
E I ++W LP+ +R KS I
Sbjct 83 ETVIQHKWKSLPQVQREGIKSYII 106
> dre:566321 importin-13-like
Length=945
Score = 48.1 bits (113), Expect = 6e-06, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 42/85 (49%), Gaps = 0/85 (0%)
Query 17 EAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFAL 76
E V ++R + ++ D +++VA + L E + P AW +L K P +FF
Sbjct 5 EFTVEAVERALQQLYYDPDMGKKNVAQKWLSEAQASPQAWQFCWDLLRPEKVPEIQFFGA 64
Query 77 QILEATITNRWNVLPETERNSTKSR 101
L A I+ W+ LP + +S +S+
Sbjct 65 STLHAKISRHWSELPAGQLDSLRSQ 89
> cpv:cgd6_3520 hypothetical protein
Length=1399
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 20/53 (37%), Positives = 31/53 (58%), Gaps = 0/53 (0%)
Query 27 VAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
+ ++ SNDS +R A++ L EFK+ AW + +L VS +P K+ A Q L
Sbjct 12 ILELWTSNDSFKRAEANKYLLEFKDSFQAWQICSELLEVSVEPEVKYVAAQTL 64
> mmu:320938 Tnpo3, 5730544L10Rik, C430013M08Rik, C81142, D6Ertd313e,
KIAA4133, MGC90049, Trn-SR, mKIAA4133; transportin 3
Length=923
Score = 31.2 bits (69), Expect = 0.77, Method: Composition-based stats.
Identities = 19/80 (23%), Positives = 37/80 (46%), Gaps = 0/80 (0%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
+ L+ + V A++ D ++ A LGE + +AW +L + +D + +FA Q +
Sbjct 8 LQLVYQAVQALYHDPDPSGKERASFWLGELQRSVHAWEISDQLLQIRQDVESCYFAAQTM 67
Query 80 EATITNRWNVLPETERNSTK 99
+ I + LP S +
Sbjct 68 KMKIQTSFYELPTDSHASLR 87
> hsa:23534 TNPO3, IPO12, MTR10A, TRN-SR, TRN-SR2, TRNSR; transportin
3
Length=859
Score = 31.2 bits (69), Expect = 0.78, Method: Composition-based stats.
Identities = 19/80 (23%), Positives = 37/80 (46%), Gaps = 0/80 (0%)
Query 20 VNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQIL 79
+ L+ + V A++ D ++ A LGE + +AW +L + +D + +FA Q +
Sbjct 8 LQLVYQAVQALYHDPDPSGKERASFWLGELQRSVHAWEISDQLLQIRQDVESCYFAAQTM 67
Query 80 EATITNRWNVLPETERNSTK 99
+ I + LP S +
Sbjct 68 KMKIQTSFYELPTDSHASLR 87
> dre:394062 tnpo3, MGC55409, zgc:55409; transportin 3
Length=923
Score = 31.2 bits (69), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 36/81 (44%), Gaps = 7/81 (8%)
Query 11 PNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPN 70
P LP L+ + V A++ D ++ A LGE + AW +L + +D
Sbjct 6 PTLP-------LVYQAVQALYHDPDPAGKERASVWLGELQRSMYAWEISDQLLQLKQDIE 58
Query 71 TKFFALQILEATITNRWNVLP 91
+ +FA Q ++ I + LP
Sbjct 59 SCYFAAQTMKMKIQTSFYELP 79
> mmu:73192 Xpot, 1110004L07Rik, 3110065H13Rik, AI452076, C79645,
EXPORTIN-T; exportin, tRNA (nuclear export receptor for
tRNAs); K14288 exportin-T
Length=962
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 33/72 (45%), Gaps = 5/72 (6%)
Query 29 AMFLSNDSVERDVAHRVLGEFKNM---PNAWTHVAFIL--NVSKDPNTKFFALQILEATI 83
A+ N + + D R L F+ + P+AW A L D + KFF Q+LE +
Sbjct 5 ALLGLNPNADSDFRQRALAYFEQLKISPDAWQVCAEALAQKTYSDDHVKFFCFQVLEHQV 64
Query 84 TNRWNVLPETER 95
+++ L ++
Sbjct 65 KYKYSELSTAQQ 76
> hsa:11260 XPOT, XPO3; exportin, tRNA (nuclear export receptor
for tRNAs); K14288 exportin-T
Length=962
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 19/72 (26%), Positives = 33/72 (45%), Gaps = 5/72 (6%)
Query 29 AMFLSNDSVERDVAHRVLGEFKNM---PNAWTHVAFIL--NVSKDPNTKFFALQILEATI 83
A+ N + + D R L F+ + P+AW A L D + KFF Q+LE +
Sbjct 5 ALLGLNPNADSDFRQRALAYFEQLKISPDAWQVCAEALAQRTYSDDHVKFFCFQVLEHQV 64
Query 84 TNRWNVLPETER 95
+++ L ++
Sbjct 65 KYKYSELTTVQQ 76
> cel:T19D12.4 hypothetical protein
Length=1028
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 4 DPRALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFK 50
D R LL N A ++ D+ V+A+F DSV D +VL K
Sbjct 79 DLRGLLKNKYTVNSFAADVQDKTVSAVFSLRDSVSADDVQKVLAGSK 125
> xla:447036 tnpo3, MGC82948; transportin 3
Length=922
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 18/75 (24%), Positives = 33/75 (44%), Gaps = 0/75 (0%)
Query 25 RVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQILEATIT 84
+ V A++ D ++ A LGE + AW +L + +D + +FA Q ++ I
Sbjct 12 QAVQALYHDPDPSGKERASLWLGELQRSVYAWEIADQLLQIHQDVESCYFAAQTMKMKIQ 71
Query 85 NRWNVLPETERNSTK 99
+ LP S +
Sbjct 72 TSFYELPSDSHVSLR 86
> tgo:TGME49_022380 exportin 7, putative
Length=1147
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 35/84 (41%), Gaps = 0/84 (0%)
Query 16 NEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFA 75
+ A V L+ + A + E++ AH+VL + P + IL S + FA
Sbjct 4 DAAQVQQLELLCQAFYGGGSKSEQNEAHQVLLPLASNPANVPRLQIILAKSNNLQALLFA 63
Query 76 LQILEATITNRWNVLPETERNSTK 99
L T W+ +P+ + T+
Sbjct 64 TAGLTNLFTKHWSQIPDPLKQDTR 87
> pfa:PFI0490c ran-binding protein, putative
Length=1198
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 1/84 (1%)
Query 16 NEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFA 75
+E+ + L + AM+ N E++ AH +L N + + IL + +T F
Sbjct 2 SESELQQLQVLCEAMYCGNKE-EQNQAHTILLPLVNNVMNVSKLKNILGSTNHVHTLIFT 60
Query 76 LQILEATITNRWNVLPETERNSTK 99
L ITN WN + + E++ K
Sbjct 61 TSGLLQLITNEWNKIDQKEKDELK 84
> cpv:cgd7_1670 nuclear pore protein RBP16/17 (RAN binding protein
16/17)
Length=1132
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 17/66 (25%), Positives = 29/66 (43%), Gaps = 0/66 (0%)
Query 35 DSVERDVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQILEATITNRWNVLPETE 94
D ++ AH VL T + +L S +P+ FA L T+ W +P+ +
Sbjct 5 DVNQQKQAHEVLLPLTCNLGCLTQLQALLAQSSNPHALMFAATGLSKLFTSCWAQIPDNQ 64
Query 95 RNSTKS 100
+ TK+
Sbjct 65 KEETKT 70
> dre:266749 pak2a, cb422, kinase, pak2, si:dz198m22.1, wu:fb71h01,
zgc:91798; p21 (CDKN1A)-activated kinase 2a (EC:2.7.1.-);
K04410 p21-activated kinase 2 [EC:2.7.11.1]
Length=517
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 15/62 (24%), Positives = 30/62 (48%), Gaps = 5/62 (8%)
Query 45 VLGEFKNMPNAWTHVAFILNVS-----KDPNTKFFALQILEATITNRWNVLPETERNSTK 99
V GEF MP W + N++ K+P L+ ++T +R L T++++ +
Sbjct 92 VTGEFTGMPEQWARLLQTSNITKSEQKKNPQAVLDVLKFYDSTGNSRQKYLSFTDKDAPQ 151
Query 100 SR 101
++
Sbjct 152 AK 153
> bbo:BBOV_I003600 19.m02390; hypothetical protein
Length=4820
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 9/32 (28%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 49 FKNMPNAWTHVAFILNVSKDPNTKFFALQILE 80
+ + ++WT ++F++N ++DP K L++L+
Sbjct 667 LRTVGDSWTRLSFVMNRNEDPAMKELFLRLLD 698
> dre:559657 zgc:101095; si:dkeyp-11g8.6
Length=835
Score = 27.7 bits (60), Expect = 9.0, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 6 RALLDPNLPFNEAAVNLLDRVVAAMFLSNDSVERDVAHRVLGEFKNMPNAWTHV 59
+ALLDP++ F+EA DRV+ + +V + + L PN H+
Sbjct 447 KALLDPSVNFSEAMQKASDRVLKQFDYDSSTVRKRIIQEALVNI-TFPNIKKHL 499
> mmu:29875 Iqgap1, AA682088, D7Ertd237e, D7Ertd257e, mKIAA0051;
IQ motif containing GTPase activating protein 1; K05767 IQ
motif containing GTPase activating protein
Length=1657
Score = 27.7 bits (60), Expect = 9.6, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 30/64 (46%), Gaps = 1/64 (1%)
Query 40 DVAHRVLGEFKNMPNAWTHVAFILNVSKDPNTKFFALQILEATITNRWNVLPETERNSTK 99
D H +L + +P + + S DPN + A + T+TN+++V P E
Sbjct 1322 DPIHELLDDLGEVPTIESLIGESCGNSNDPNKEALAKTEVSLTLTNKFDV-PGDENAEMD 1380
Query 100 SRSI 103
+R+I
Sbjct 1381 ARTI 1384
Lambda K H
0.320 0.132 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2069971060
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40