bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2023_orf1
Length=81
Score E
Sequences producing significant alignments: (Bits) Value
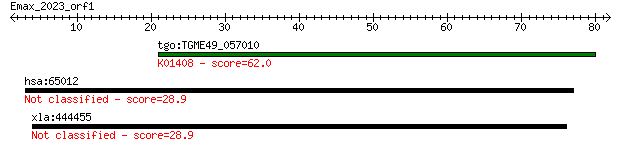
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 62.0 4e-10
hsa:65012 SLC26A10, FLJ32854; solute carrier family 26, member... 28.9 3.7
xla:444455 scfd1, MGC83661; sec1 family domain containing 1 28.9
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 27/59 (45%), Positives = 38/59 (64%), Gaps = 0/59 (0%)
Query 21 LAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLMEHVAKLLSDPSM 79
+AA+AE L EETVD + CG++HS+ G G + F AYT QL ++M VA + DP +
Sbjct 580 MAALAEQLDEETVDLKQCGISHSVGVSGDGLLLAFAAYTPKQLRQVMAVVASKIQDPQV 638
> hsa:65012 SLC26A10, FLJ32854; solute carrier family 26, member
10; K14707 solute carrier family 26, member 10
Length=563
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 36/86 (41%), Gaps = 12/86 (13%)
Query 3 GWQRHLAPAT-----------IQRSLRFPLAAIAEHLQEETVDFQNCGVTHSLAFKGTGF 51
G RHL+ T ++R + PL +++E +D Q GV ++AF
Sbjct 70 GTGRHLSTGTFAILSLMTGSAVERLVPEPLVGNLSGIEKEQLDAQRVGVAAAVAFGSGAL 129
Query 52 HMGFEAYTEGQLSKLM-EHVAKLLSD 76
+G G LS + E V K L+
Sbjct 130 MLGMFVLQLGVLSTFLSEPVVKALTS 155
> xla:444455 scfd1, MGC83661; sec1 family domain containing 1
Length=632
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 30/72 (41%), Gaps = 13/72 (18%)
Query 4 WQRHLAPATIQRSLRFPLAAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQL 63
WQ+H P +AE +Q+E ++ T K MG E EG +
Sbjct 316 WQKHKGS---------PFPEVAESVQQELESYR----TQEDEVKRLKTIMGLEGEDEGAI 362
Query 64 SKLMEHVAKLLS 75
S + ++ AKL S
Sbjct 363 SMISDNTAKLTS 374
Lambda K H
0.320 0.132 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2053886800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40