bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2116_orf3
Length=95
Score E
Sequences producing significant alignments: (Bits) Value
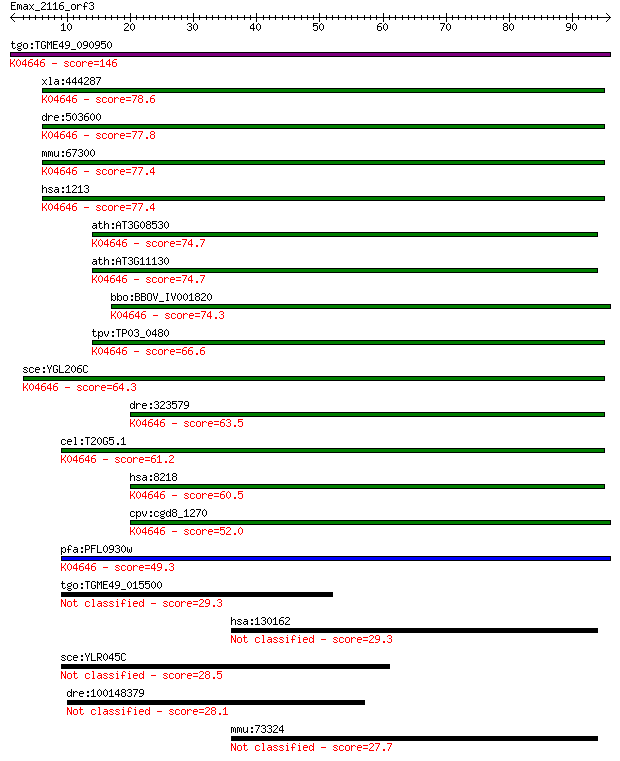
tgo:TGME49_090950 clathrin heavy chain, putative ; K04646 clat... 146 2e-35
xla:444287 cltc, MGC80936; clathrin, heavy chain (Hc); K04646 ... 78.6 4e-15
dre:503600 cltcb, im:7145213, zgc:113234; clathrin, heavy poly... 77.8 8e-15
mmu:67300 Cltc, 3110065L21Rik, CHC, MGC92975, R74732; clathrin... 77.4 9e-15
hsa:1213 CLTC, CHC, CHC17, CLH-17, CLTCL2, Hc, KIAA0034; clath... 77.4 9e-15
ath:AT3G08530 clathrin heavy chain, putative; K04646 clathrin ... 74.7 7e-14
ath:AT3G11130 clathrin heavy chain, putative; K04646 clathrin ... 74.7 7e-14
bbo:BBOV_IV001820 21.m02773; clathrin heavy chain; K04646 clat... 74.3 8e-14
tpv:TP03_0480 clathrin heavy chain; K04646 clathrin heavy chain 66.6 2e-11
sce:YGL206C CHC1, SWA5; Chc1p; K04646 clathrin heavy chain 64.3 9e-11
dre:323579 cltca, cb1033, cltc, wu:fc03e11, wu:fc49a11, wu:fd0... 63.5 1e-10
cel:T20G5.1 chc-1; Clathrin Heavy Chain family member (chc-1);... 61.2 9e-10
hsa:8218 CLTCL1, CHC22, CLH22, CLTCL, CLTD, FLJ36032; clathrin... 60.5 1e-09
cpv:cgd8_1270 clathrin heavy chain ; K04646 clathrin heavy chain 52.0 5e-07
pfa:PFL0930w clathrin heavy chain, putative; K04646 clathrin h... 49.3 3e-06
tgo:TGME49_015500 hypothetical protein 29.3 2.9
hsa:130162 C2orf63, FLJ31438; chromosome 2 open reading frame 63 29.3
sce:YLR045C STU2; Microtubule-associated protein (MAP) of the ... 28.5 6.0
dre:100148379 MGC113421, wu:fc49c05, wu:fi39c11; zgc:113421 28.1 7.3
mmu:73324 1700034F02Rik; RIKEN cDNA 1700034F02 gene 27.7 9.7
> tgo:TGME49_090950 clathrin heavy chain, putative ; K04646 clathrin
heavy chain
Length=1731
Score = 146 bits (368), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 70/95 (73%), Positives = 81/95 (85%), Gaps = 0/95 (0%)
Query 1 LGPYMQHALPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKS 60
L PY+ AL ++PNR QI SLAKRYGLPGA++TL+Q FNQHFASG+++ AARI ATLKS
Sbjct 360 LIPYINQALVYVPNRQQIATSLAKRYGLPGAEETLMQEFNQHFASGNFKTAARIAATLKS 419
Query 61 GRLRTPQVLQQFKGVPAQPGQTSAILLYFSTLLEY 95
G LRT QV+QQFK VP QPGQTSAIL+YFSTLLEY
Sbjct 420 GVLRTAQVIQQFKSVPTQPGQTSAILVYFSTLLEY 454
> xla:444287 cltc, MGC80936; clathrin, heavy chain (Hc); K04646
clathrin heavy chain
Length=1675
Score = 78.6 bits (192), Expect = 4e-15, Method: Composition-based stats.
Identities = 38/92 (41%), Positives = 58/92 (63%), Gaps = 3/92 (3%)
Query 6 QHALPHLPNRAQ---IYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
++ +P++ N Q + L +A R L GA++ + FN FA G+Y AA++ A G
Sbjct 332 ENIIPYITNVLQNPDLALRMAVRNNLAGAEELFARKFNALFAQGNYSEAAKVAANAPKGI 391
Query 63 LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
LRTP+ +++F+ VPAQPGQTS +L YF LL+
Sbjct 392 LRTPETIRRFQSVPAQPGQTSPLLQYFGILLD 423
> dre:503600 cltcb, im:7145213, zgc:113234; clathrin, heavy polypeptide
b (Hc); K04646 clathrin heavy chain
Length=1677
Score = 77.8 bits (190), Expect = 8e-15, Method: Composition-based stats.
Identities = 38/92 (41%), Positives = 57/92 (61%), Gaps = 3/92 (3%)
Query 6 QHALPHLPNRAQ---IYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
++ +P++ N Q + L +A R L GA++ + FN FA G+Y AA++ A G
Sbjct 332 ENIIPYITNVLQNPDLALRMAVRNNLAGAEELFARKFNNLFAGGNYSEAAKVAANAPKGI 391
Query 63 LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
LRTP +++F+ VPAQPGQTS +L YF LL+
Sbjct 392 LRTPDTIRRFQSVPAQPGQTSPLLQYFGILLD 423
> mmu:67300 Cltc, 3110065L21Rik, CHC, MGC92975, R74732; clathrin,
heavy polypeptide (Hc); K04646 clathrin heavy chain
Length=1675
Score = 77.4 bits (189), Expect = 9e-15, Method: Composition-based stats.
Identities = 38/92 (41%), Positives = 57/92 (61%), Gaps = 3/92 (3%)
Query 6 QHALPHLPNRAQ---IYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
++ +P++ N Q + L +A R L GA++ + FN FA G+Y AA++ A G
Sbjct 332 ENIIPYITNVLQNPDLALRMAVRNNLAGAEELFARKFNALFAQGNYSEAAKVAANAPKGI 391
Query 63 LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
LRTP +++F+ VPAQPGQTS +L YF LL+
Sbjct 392 LRTPDTIRRFQSVPAQPGQTSPLLQYFGILLD 423
> hsa:1213 CLTC, CHC, CHC17, CLH-17, CLTCL2, Hc, KIAA0034; clathrin,
heavy chain (Hc); K04646 clathrin heavy chain
Length=1675
Score = 77.4 bits (189), Expect = 9e-15, Method: Composition-based stats.
Identities = 38/92 (41%), Positives = 57/92 (61%), Gaps = 3/92 (3%)
Query 6 QHALPHLPNRAQ---IYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
++ +P++ N Q + L +A R L GA++ + FN FA G+Y AA++ A G
Sbjct 332 ENIIPYITNVLQNPDLALRMAVRNNLAGAEELFARKFNALFAQGNYSEAAKVAANAPKGI 391
Query 63 LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
LRTP +++F+ VPAQPGQTS +L YF LL+
Sbjct 392 LRTPDTIRRFQSVPAQPGQTSPLLQYFGILLD 423
> ath:AT3G08530 clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1703
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 36/80 (45%), Positives = 50/80 (62%), Gaps = 0/80 (0%)
Query 14 NRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFK 73
N ++ ++LAKR LPGA++ +VQ F + FA Y+ AA + A G LRTP + +F+
Sbjct 356 NNLELAVNLAKRGNLPGAENLVVQRFQELFAQTKYKEAAELAAESPQGILRTPDTVAKFQ 415
Query 74 GVPAQPGQTSAILLYFSTLL 93
VP Q GQT +L YF TLL
Sbjct 416 SVPVQAGQTPPLLQYFGTLL 435
> ath:AT3G11130 clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1705
Score = 74.7 bits (182), Expect = 7e-14, Method: Composition-based stats.
Identities = 36/80 (45%), Positives = 50/80 (62%), Gaps = 0/80 (0%)
Query 14 NRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFK 73
N ++ ++LAKR LPGA++ +VQ F + FA Y+ AA + A G LRTP + +F+
Sbjct 356 NNLELAVNLAKRGNLPGAENLVVQRFQELFAQTKYKEAAELAAESPQGILRTPDTVAKFQ 415
Query 74 GVPAQPGQTSAILLYFSTLL 93
VP Q GQT +L YF TLL
Sbjct 416 SVPVQAGQTPPLLQYFGTLL 435
> bbo:BBOV_IV001820 21.m02773; clathrin heavy chain; K04646 clathrin
heavy chain
Length=1676
Score = 74.3 bits (181), Expect = 8e-14, Method: Composition-based stats.
Identities = 31/79 (39%), Positives = 55/79 (69%), Gaps = 0/79 (0%)
Query 17 QIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVP 76
++ +SLA R+G PG++ ++++F ++F+ +Y+ AA + ATLK+G LRT + +++F P
Sbjct 356 EVRISLATRFGYPGSESLMMRSFEEYFSRREYKQAALLVATLKNGALRTNETMERFLNAP 415
Query 77 AQPGQTSAILLYFSTLLEY 95
G+TS L YFS +LE+
Sbjct 416 VLAGETSPALHYFSVMLEH 434
> tpv:TP03_0480 clathrin heavy chain; K04646 clathrin heavy chain
Length=1696
Score = 66.6 bits (161), Expect = 2e-11, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 51/81 (62%), Gaps = 2/81 (2%)
Query 14 NRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFK 73
N ++ + +A YGL G+DD L+++F F + YR AA+I ATLKS +LRT +Q+FK
Sbjct 371 NLKEVSVGIATCYGLKGSDDLLLESFESFFNNEQYRQAAQIVATLKSNQLRTLDTIQRFK 430
Query 74 GVPAQPGQTSAILLYFSTLLE 94
+ G +A+ YFS LLE
Sbjct 431 NKTTETG--NALSHYFSVLLE 449
> sce:YGL206C CHC1, SWA5; Chc1p; K04646 clathrin heavy chain
Length=1653
Score = 64.3 bits (155), Expect = 9e-11, Method: Composition-based stats.
Identities = 36/92 (39%), Positives = 51/92 (55%), Gaps = 6/92 (6%)
Query 3 PYMQHALPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR 62
PY+ + L ++ + L +A R GLPGADD + F DY+ AA++ A+ S
Sbjct 342 PYILNKLSNVA----LALIVATRGGLPGADDLFQKQFESLLLQNDYQNAAKVAAS--STS 395
Query 63 LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
LR + + K + A PG S ILLYFSTLL+
Sbjct 396 LRNQNTINRLKNIQAPPGAISPILLYFSTLLD 427
> dre:323579 cltca, cb1033, cltc, wu:fc03e11, wu:fc49a11, wu:fd07f02;
clathrin, heavy polypeptide a (Hc); K04646 clathrin
heavy chain
Length=1680
Score = 63.5 bits (153), Expect = 1e-10, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 48/75 (64%), Gaps = 0/75 (0%)
Query 20 LSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQP 79
L +A R L GA++ + FN FA G++ AA++ A G LRTP +++F+ VPAQP
Sbjct 349 LRMAVRNNLAGAEELFGRKFNNLFAGGNFAEAAKVAANAPKGILRTPDTIRRFQSVPAQP 408
Query 80 GQTSAILLYFSTLLE 94
GQTS +L YF LL+
Sbjct 409 GQTSPLLQYFGILLD 423
> cel:T20G5.1 chc-1; Clathrin Heavy Chain family member (chc-1);
K04646 clathrin heavy chain
Length=1681
Score = 61.2 bits (147), Expect = 9e-10, Method: Composition-based stats.
Identities = 32/89 (35%), Positives = 52/89 (58%), Gaps = 3/89 (3%)
Query 9 LPHLPNRAQ---IYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRT 65
+P + N+ Q + L LA R LPGA++ V+ FN F++G + +A++ A+ G LRT
Sbjct 333 VPFVTNQLQNPDLALKLAVRCDLPGAEELFVRKFNLLFSNGQFGESAKVAASAPQGILRT 392
Query 66 PQVLQQFKGVPAQPGQTSAILLYFSTLLE 94
P +Q+F+ P+ S +L YF LL+
Sbjct 393 PATIQKFQQCPSTGPGPSPLLQYFGILLD 421
> hsa:8218 CLTCL1, CHC22, CLH22, CLTCL, CLTD, FLJ36032; clathrin,
heavy chain-like 1; K04646 clathrin heavy chain
Length=1583
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 34/75 (45%), Positives = 46/75 (61%), Gaps = 0/75 (0%)
Query 20 LSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQP 79
L LA R L GA+ V+ FN FA G Y AA++ A+ G LRT + +Q+F+ +PAQ
Sbjct 349 LRLAVRSNLAGAEKLFVRKFNTLFAQGSYAEAAKVAASAPKGILRTRETVQKFQSIPAQS 408
Query 80 GQTSAILLYFSTLLE 94
GQ S +L YF LL+
Sbjct 409 GQASPLLQYFGILLD 423
> cpv:cgd8_1270 clathrin heavy chain ; K04646 clathrin heavy chain
Length=2007
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 25/76 (32%), Positives = 40/76 (52%), Gaps = 3/76 (3%)
Query 20 LSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQP 79
L +RYG G D+ ++ FN+ DY+ A R+ + K+G LRTP L FK V +
Sbjct 386 LRWTQRYGYQGTDEFSIRMFNESIKRQDYQNACRVVSLNKNGSLRTPATLNHFKMVDS-- 443
Query 80 GQTSAILLYFSTLLEY 95
+ YF+T+ ++
Sbjct 444 -SNKLLFQYFTTVFKF 458
> pfa:PFL0930w clathrin heavy chain, putative; K04646 clathrin
heavy chain
Length=1997
Score = 49.3 bits (116), Expect = 3e-06, Method: Composition-based stats.
Identities = 34/115 (29%), Positives = 50/115 (43%), Gaps = 30/115 (26%)
Query 9 LPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAARICATLKSGR------ 62
L +L + +I +L +YG PG D + A+ + D++ A++I +K+ +
Sbjct 378 LSNLEFKDKIIKNLCVKYGYPGCD--YISAYKKCINDMDFKKASKIICLMKNTKIFEEHS 435
Query 63 ----------------------LRTPQVLQQFKGVPAQPGQTSAILLYFSTLLEY 95
LRT QVL FK GQ S +LLYFS LLEY
Sbjct 436 NNIAESILIMNRKNIDIRIIPPLRTQQVLNSFKSFKNTSGQLSPLLLYFSVLLEY 490
> tgo:TGME49_015500 hypothetical protein
Length=574
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 17/43 (39%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 9 LPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGA 51
L HL N+AQ L L K A T++Q ++H A YR A
Sbjct 257 LTHLRNQAQTALKLEKFQEAIKACTTIIQDIDEHDAKARYRRA 299
> hsa:130162 C2orf63, FLJ31438; chromosome 2 open reading frame
63
Length=464
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 26/58 (44%), Gaps = 0/58 (0%)
Query 36 VQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQPGQTSAILLYFSTLL 93
++ FN+ + G+Y AA A LR + FK V G+ +LL+F L
Sbjct 176 IERFNELISLGEYEKAACYAANSPRRILRNIGTMNTFKAVGKIRGKPLPLLLFFEALF 233
> sce:YLR045C STU2; Microtubule-associated protein (MAP) of the
XMAP215/Dis1 family; regulates microtubule dynamics during
spindle orientation and metaphase chromosome alignment; interacts
with spindle pole body component Spc72p
Length=888
Score = 28.5 bits (62), Expect = 6.0, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 25/53 (47%), Gaps = 1/53 (1%)
Query 9 LPHLPNRAQIYLSLAKRYGLPGADDTLVQAFNQHFASGD-YRGAARICATLKS 60
LPH N I G+ D L FN FAS + Y+ AA + +TLK+
Sbjct 819 LPHRVNSLNIRPYRKNGTGVSSVSDDLDIDFNDSFASEESYKRAAAVTSTLKA 871
> dre:100148379 MGC113421, wu:fc49c05, wu:fi39c11; zgc:113421
Length=1912
Score = 28.1 bits (61), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 26/55 (47%), Gaps = 8/55 (14%)
Query 10 PHLPNR-----AQIYLSLAKRYGLPGADDTLVQAFNQHFASGDYRGAAR---ICA 56
PH+PN+ A Y LAK + ++ F Q ASG ++G R +CA
Sbjct 1304 PHVPNQPSEAAAHFYFELAKTVLIKAGGNSSTSIFTQPSASGGHQGPHRNLHLCA 1358
> mmu:73324 1700034F02Rik; RIKEN cDNA 1700034F02 gene
Length=585
Score = 27.7 bits (60), Expect = 9.7, Method: Composition-based stats.
Identities = 14/58 (24%), Positives = 26/58 (44%), Gaps = 0/58 (0%)
Query 36 VQAFNQHFASGDYRGAARICATLKSGRLRTPQVLQQFKGVPAQPGQTSAILLYFSTLL 93
++ FN + G+Y AA A L+ + +FK + G+ +LL+F +
Sbjct 298 IERFNDLISLGEYERAACFAANSPKRILQNTSTMNKFKAIGKIRGKPLPLLLFFEAIF 355
Lambda K H
0.323 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2065224908
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40