bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2187_orf1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
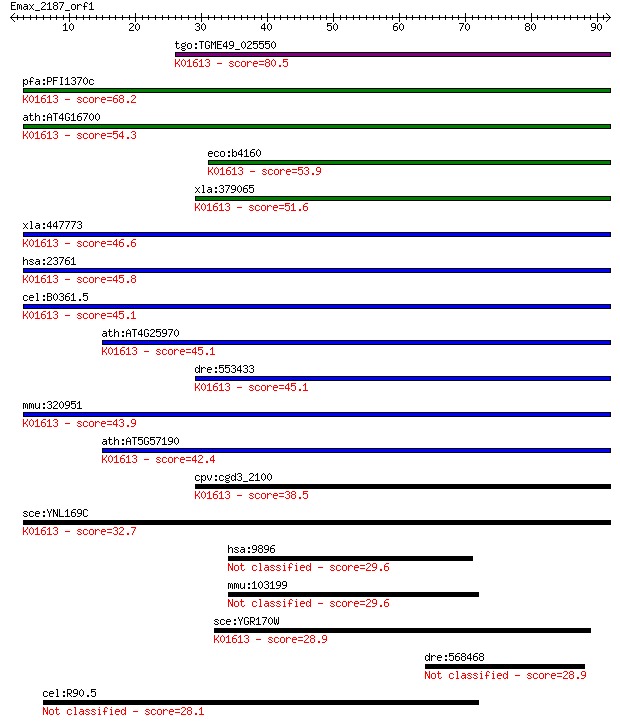
tgo:TGME49_025550 phosphatidylserine decarboxylase proenzyme, ... 80.5 1e-15
pfa:PFI1370c PfPSD; phosphatidylserine decarboxylase (EC:4.1.1... 68.2 7e-12
ath:AT4G16700 PSD1; PSD1 (phosphatidylserine decarboxylase 1);... 54.3 1e-07
eco:b4160 psd, ECK4156, JW4121; phosphatidylserine decarboxyla... 53.9 1e-07
xla:379065 MGC52759; similar to phosphatidylserine decarboxyla... 51.6 6e-07
xla:447773 pisd, MGC84353; phosphatidylserine decarboxylase (E... 46.6 2e-05
hsa:23761 PISD, DJ858B16, DKFZp566G2246, PSD, PSDC, PSSC, dJ85... 45.8 4e-05
cel:B0361.5 psd-1; PhosphatidylSerine Decarboxylase family mem... 45.1 5e-05
ath:AT4G25970 PSD3; PSD3 (phosphatidylserine decarboxylase 3);... 45.1 5e-05
dre:553433 pisd, wu:fd05a08, zgc:158135; phosphatidylserine de... 45.1 6e-05
mmu:320951 Pisd, 9030221M09Rik; phosphatidylserine decarboxyla... 43.9 1e-04
ath:AT5G57190 PSD2; PSD2 (phosphatidylserine decarboxylase 2);... 42.4 4e-04
cpv:cgd3_2100 phosphatidylserine decarboxylase ; K01613 phosph... 38.5 0.006
sce:YNL169C PSD1; Phosphatidylserine decarboxylase of the mito... 32.7 0.28
hsa:9896 FIG4, ALS11, CMT4J, KIAA0274, SAC3, dJ249I4.1; FIG4 h... 29.6 2.2
mmu:103199 Fig4, A530089I17Rik, AI326867, Sac3; FIG4 homolog (... 29.6 2.6
sce:YGR170W PSD2; Phosphatidylserine decarboxylase of the Golg... 28.9 3.9
dre:568468 fat2; FAT tumor suppressor homolog 2 (Drosophila) 28.9 4.2
cel:R90.5 glb-24; GLoBin family member (glb-24) 28.1 7.4
> tgo:TGME49_025550 phosphatidylserine decarboxylase proenzyme,
putative (EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=337
Score = 80.5 bits (197), Expect = 1e-15, Method: Composition-based stats.
Identities = 37/66 (56%), Positives = 46/66 (69%), Gaps = 0/66 (0%)
Query 26 RYPLEAYESFGDLFCRTLRDKAREIMDLSPFAMVSPCDCTVSILGTVEGDRVPQVKGATY 85
RYPL +Y+ G LF RTL+DK REI D+ ++ SP D V+ LG V +RV QVKGATY
Sbjct 83 RYPLRSYKCIGHLFARTLKDKEREIEDIGTQSLASPADGVVTALGDVSSERVEQVKGATY 142
Query 86 SLKGFL 91
SL+ FL
Sbjct 143 SLRAFL 148
> pfa:PFI1370c PfPSD; phosphatidylserine decarboxylase (EC:4.1.1.65);
K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=353
Score = 68.2 bits (165), Expect = 7e-12, Method: Composition-based stats.
Identities = 32/90 (35%), Positives = 54/90 (60%), Gaps = 1/90 (1%)
Query 3 IPVSLRPLLMSIFL-HLQPAAADSRYPLEAYESFGDLFCRTLRDKAREIMDLSPFAMVSP 61
IP S R + + F+ +L + +YP+E+Y+S GD F R +R+ R I DL+ +++VSP
Sbjct 77 IPHSYRLYVYNFFIKYLNINKEEIKYPIESYKSLGDFFSRYIREDTRPIGDLNEYSIVSP 136
Query 62 CDCTVSILGTVEGDRVPQVKGATYSLKGFL 91
CD + G + + + VKG +++K FL
Sbjct 137 CDSEIVDFGELTSNYLDNVKGIKFNIKTFL 166
> ath:AT4G16700 PSD1; PSD1 (phosphatidylserine decarboxylase 1);
phosphatidylserine decarboxylase (EC:4.1.1.65); K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=453
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 34/92 (36%), Positives = 48/92 (52%), Gaps = 4/92 (4%)
Query 3 IPVSLRPLLMSIFLH-LQPAAADSRYPLEAYESFGDLFCRTLRDKAREIMDLSPFAMVSP 61
IPV +RP + ++ PLE Y S D F R+L++ R I D P +VSP
Sbjct 139 IPVWMRPYAYKAWARAFHSNLEEAALPLEEYTSLQDFFVRSLKEGCRPI-DPDPCCLVSP 197
Query 62 CDCTVSILGTVEGDR--VPQVKGATYSLKGFL 91
D TV G ++G+R + QVKG +YS+ L
Sbjct 198 VDGTVLRFGELKGNRGMIEQVKGHSYSVPALL 229
> eco:b4160 psd, ECK4156, JW4121; phosphatidylserine decarboxylase
(EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=322
Score = 53.9 bits (128), Expect = 1e-07, Method: Composition-based stats.
Identities = 26/61 (42%), Positives = 36/61 (59%), Gaps = 1/61 (1%)
Query 31 AYESFGDLFCRTLRDKAREIMDLSPFAMVSPCDCTVSILGTVEGDRVPQVKGATYSLKGF 90
+Y +F + F R LRD+ R I D P +V P D +S LG +E D++ Q KG YSL+
Sbjct 59 SYRTFNEFFVRPLRDEVRPI-DTDPNVLVMPADGVISQLGKIEEDKILQAKGHNYSLEAL 117
Query 91 L 91
L
Sbjct 118 L 118
> xla:379065 MGC52759; similar to phosphatidylserine decarboxylase;
K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=355
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 37/63 (58%), Gaps = 3/63 (4%)
Query 29 LEAYESFGDLFCRTLRDKAREIMDLSPFAMVSPCDCTVSILGTVEGDRVPQVKGATYSLK 88
L Y++ G+LF R+L+ AR I +VSPCD V G G ++ QVKG TYSL+
Sbjct 111 LNRYQNLGELFRRSLKPTARTI---DSHCVVSPCDGRVLHSGRGRGSQIEQVKGLTYSLE 167
Query 89 GFL 91
FL
Sbjct 168 SFL 170
> xla:447773 pisd, MGC84353; phosphatidylserine decarboxylase
(EC:4.1.1.65); K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=411
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 50/94 (53%), Gaps = 10/94 (10%)
Query 3 IPVSLRPLLMSIFL-----HLQPAAADSRYPLEAYESFGDLFCRTLRDKAREIMDLSPFA 57
+P LR + S+++ +++ AA + L Y + + F R L+ +AR + D +
Sbjct 138 LPTWLRKPVYSLYIWTFGVNMKEAAVED---LHQYRNLSEFFRRKLKPQARPVCD--SHS 192
Query 58 MVSPCDCTVSILGTVEGDRVPQVKGATYSLKGFL 91
++SP D + G V+ V QVKG TYSL+ FL
Sbjct 193 VISPSDGKILHFGRVKNCEVEQVKGVTYSLESFL 226
> hsa:23761 PISD, DJ858B16, DKFZp566G2246, PSD, PSDC, PSSC, dJ858B16.2;
phosphatidylserine decarboxylase (EC:4.1.1.65); K01613
phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=375
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/94 (31%), Positives = 50/94 (53%), Gaps = 10/94 (10%)
Query 3 IPVSLRPLLMSIFL-----HLQPAAADSRYPLEAYESFGDLFCRTLRDKAREIMDLSPFA 57
+P LR + S+++ +++ AA + L Y + + F R L+ +AR + L +
Sbjct 97 LPHWLRRPVYSLYIWTFGVNMKEAAVED---LHHYRNLSEFFRRKLKPQARPVCGLH--S 151
Query 58 MVSPCDCTVSILGTVEGDRVPQVKGATYSLKGFL 91
++SP D + G V+ V QVKG TYSL+ FL
Sbjct 152 VISPSDGRILNFGQVKNCEVEQVKGVTYSLESFL 185
> cel:B0361.5 psd-1; PhosphatidylSerine Decarboxylase family member
(psd-1); K01613 phosphatidylserine decarboxylase [EC:4.1.1.65]
Length=377
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 32/91 (35%), Positives = 42/91 (46%), Gaps = 5/91 (5%)
Query 3 IPVSLRPLLMSIFLHLQPAAADS--RYPLEAYESFGDLFCRTLRDKAREIMDLSPFAMVS 60
IPV LR L+ F + D + Y SF F R L++ R I S +VS
Sbjct 122 IPVWLREHLLGGFARMYDCRMDDCVDPDFKNYPSFAAFFNRKLKESTRPI---SASPLVS 178
Query 61 PCDCTVSILGTVEGDRVPQVKGATYSLKGFL 91
P D TV G VE +++ VKG Y + FL
Sbjct 179 PADGTVLHFGKVEDNKIEYVKGHDYDVDKFL 209
> ath:AT4G25970 PSD3; PSD3 (phosphatidylserine decarboxylase 3);
phosphatidylserine decarboxylase (EC:4.1.1.65); K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=635
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 41/79 (51%), Gaps = 3/79 (3%)
Query 15 FLHLQPAAADSRYPLEAYESFGDLFCRTLRDKAREI--MDLSPFAMVSPCDCTVSILGTV 72
F Q A+ +YPL+ +++F + F R L+ AR I MD A VS DC + +V
Sbjct 393 FFKDQINMAEVKYPLDHFKTFNEFFVRELKPGARPIACMDQDDVA-VSAADCRLMAFQSV 451
Query 73 EGDRVPQVKGATYSLKGFL 91
+ +KG +S+KG L
Sbjct 452 DDSTRFWIKGRKFSIKGLL 470
> dre:553433 pisd, wu:fd05a08, zgc:158135; phosphatidylserine
decarboxylase (EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=426
Score = 45.1 bits (105), Expect = 6e-05, Method: Composition-based stats.
Identities = 24/63 (38%), Positives = 35/63 (55%), Gaps = 2/63 (3%)
Query 29 LEAYESFGDLFCRTLRDKAREIMDLSPFAMVSPCDCTVSILGTVEGDRVPQVKGATYSLK 88
L+ Y + G+ F R L+ + R + D ++SP D + G V+ V QVKG TYSL+
Sbjct 175 LQHYRNLGEFFRRKLKPQVRPVCD--SHCVISPADGKILHFGRVKNCEVEQVKGVTYSLE 232
Query 89 GFL 91
FL
Sbjct 233 TFL 235
> mmu:320951 Pisd, 9030221M09Rik; phosphatidylserine decarboxylase
(EC:4.1.1.65); K01613 phosphatidylserine decarboxylase
[EC:4.1.1.65]
Length=406
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 30/94 (31%), Positives = 47/94 (50%), Gaps = 10/94 (10%)
Query 3 IPVSLRPLLMSIFL-----HLQPAAADSRYPLEAYESFGDLFCRTLRDKAREIMDLSPFA 57
+P LR + S+++ ++ AA + L Y + + F R L+ +AR + L
Sbjct 128 LPYWLRRPVYSLYIWTFGVNMTEAAVED---LHHYRNLSEFFRRKLKPQARPVCGLH--C 182
Query 58 MVSPCDCTVSILGTVEGDRVPQVKGATYSLKGFL 91
+ SP D + G V+ V QVKG TYSL+ FL
Sbjct 183 VTSPSDGKILTFGQVKNSEVEQVKGVTYSLESFL 216
> ath:AT5G57190 PSD2; PSD2 (phosphatidylserine decarboxylase 2);
phosphatidylserine decarboxylase (EC:4.1.1.65); K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=635
Score = 42.4 bits (98), Expect = 4e-04, Method: Composition-based stats.
Identities = 25/79 (31%), Positives = 41/79 (51%), Gaps = 3/79 (3%)
Query 15 FLHLQPAAADSRYPLEAYESFGDLFCRTLRDKAREI--MDLSPFAMVSPCDCTVSILGTV 72
F Q A+ +YPL+ +++F + F R L+ AR I M+ + A V DC + +V
Sbjct 394 FFKDQINMAEVKYPLQHFKTFNEFFIRELKPGARPIACMNRNDVA-VCAADCRLMAFQSV 452
Query 73 EGDRVPQVKGATYSLKGFL 91
E +KG +S++G L
Sbjct 453 EDSTRFWIKGKKFSIRGLL 471
> cpv:cgd3_2100 phosphatidylserine decarboxylase ; K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=314
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 33/64 (51%), Gaps = 1/64 (1%)
Query 29 LEAYESFGDLFCRTLRDKAREIMDLS-PFAMVSPCDCTVSILGTVEGDRVPQVKGATYSL 87
L++Y S G+LF R++R L P ++ SPC+ G + D+ QVK +T+ +
Sbjct 67 LDSYRSIGELFTRSIRPSEIVFQCLEDPNSISSPCEGRTIEFGEINSDKCIQVKSSTFKV 126
Query 88 KGFL 91
L
Sbjct 127 SELL 130
> sce:YNL169C PSD1; Phosphatidylserine decarboxylase of the mitochondrial
inner membrane, converts phosphatidylserine to phosphatidylethanolamine
(EC:4.1.1.65); K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=500
Score = 32.7 bits (73), Expect = 0.28, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 41/93 (44%), Gaps = 5/93 (5%)
Query 3 IPVSLRPLLMSIFLHLQPAAADSRYP--LEAYESFGDLFCRTLRDKAREIMDLSPFAMVS 60
+P+ +RP ++ L D L Y + + F R ++ R + + S
Sbjct 149 LPIWVRPWGYRLYSFLFGVNLDEMEDPDLTHYANLSEFFYRNIKPGTRPVAQ-GEDVIAS 207
Query 61 PCDCTVSILGTV--EGDRVPQVKGATYSLKGFL 91
P D + +G + E + QVKG TYS+K FL
Sbjct 208 PSDGKILQVGIINSETGEIEQVKGMTYSIKEFL 240
> hsa:9896 FIG4, ALS11, CMT4J, KIAA0274, SAC3, dJ249I4.1; FIG4
homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae)
Length=907
Score = 29.6 bits (65), Expect = 2.2, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 6/43 (13%)
Query 34 SFGDLFCRTLRDKAREIM------DLSPFAMVSPCDCTVSILG 70
SF D FC + AR+ M D SPF + P + S+LG
Sbjct 685 SFDDTFCLAMTSSARDFMPKTVGIDPSPFTVRKPDETGKSVLG 727
> mmu:103199 Fig4, A530089I17Rik, AI326867, Sac3; FIG4 homolog
(S. cerevisiae)
Length=907
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 6/44 (13%)
Query 34 SFGDLFCRTLRDKAREIM------DLSPFAMVSPCDCTVSILGT 71
SF D FC + AR+ M D SPF + P + S+LG
Sbjct 685 SFDDTFCLAMTSSARDFMPKTVGIDPSPFTVRKPDETGKSVLGN 728
> sce:YGR170W PSD2; Phosphatidylserine decarboxylase of the Golgi
and vacuolar membranes, converts phosphatidylserine to phosphatidylethanolamine
(EC:4.1.1.65); K01613 phosphatidylserine
decarboxylase [EC:4.1.1.65]
Length=1138
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 14/57 (24%), Positives = 29/57 (50%), Gaps = 0/57 (0%)
Query 32 YESFGDLFCRTLRDKAREIMDLSPFAMVSPCDCTVSILGTVEGDRVPQVKGATYSLK 88
+++F + F R L+ +R + + SP D ++ T++ + VKG +S+K
Sbjct 868 FKTFNEFFYRKLKPGSRLPESNNKEILFSPADSRCTVFPTIQESKEIWVKGRKFSIK 924
> dre:568468 fat2; FAT tumor suppressor homolog 2 (Drosophila)
Length=4342
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 64 CTVSILGTVEGDRVPQVKGATYSL 87
CTV I+ T E D VPQ K + Y +
Sbjct 2562 CTVKIILTDENDNVPQFKASEYDI 2585
> cel:R90.5 glb-24; GLoBin family member (glb-24)
Length=322
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 24/77 (31%), Positives = 30/77 (38%), Gaps = 11/77 (14%)
Query 6 SLRPLLMSIFLHLQ---------PAAADSRYPLEAYESFGDLFC--RTLRDKAREIMDLS 54
SL PL + HL P A R PL + S GDL C R E++ LS
Sbjct 85 SLNPLAHTTCPHLGQMGRSRTECPEAESLRLPLTSKRSAGDLSCPPSPTRRNLNEVIGLS 144
Query 55 PFAMVSPCDCTVSILGT 71
P+ C +I T
Sbjct 145 PYQQKLLVQCWPNIYTT 161
Lambda K H
0.326 0.140 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007827920
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40