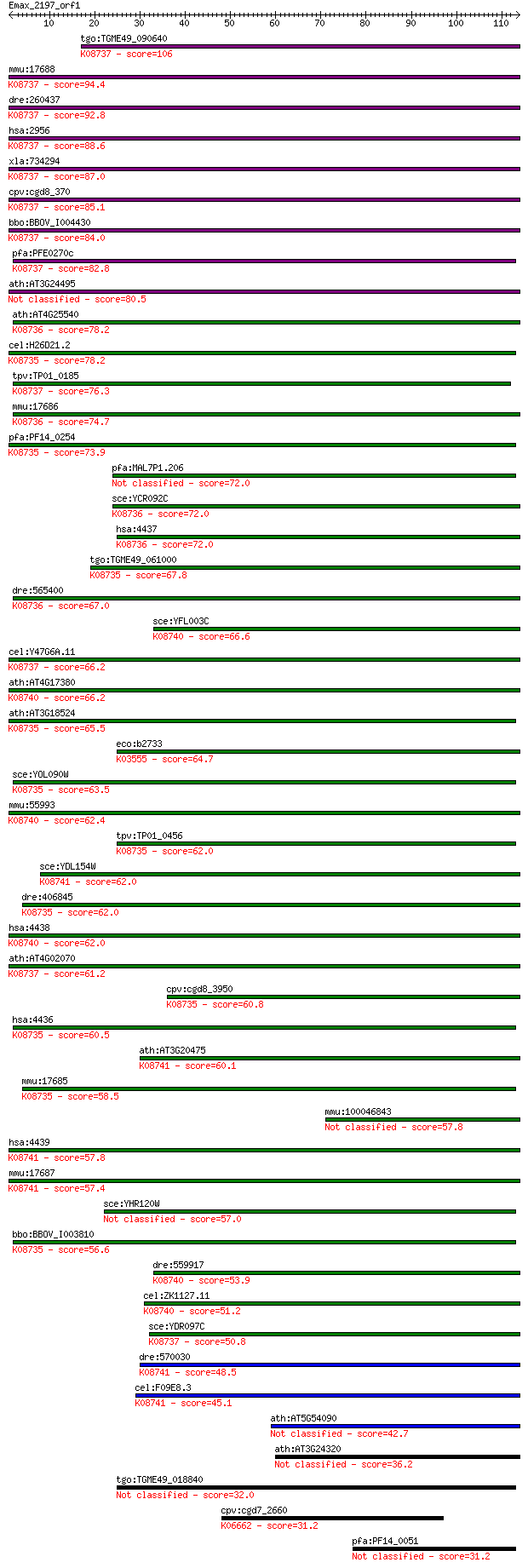
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2197_orf1
Length=113
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_090640 DNA mismatch repair protein, putative (EC:3.... 106 2e-23
mmu:17688 Msh6, AU044881, AW550279, GTBP, Gtmbp; mutS homolog ... 94.4 9e-20
dre:260437 msh6, cb326, wu:fb36c12; mutS homolog 6 (E. coli); ... 92.8 3e-19
hsa:2956 MSH6, GTBP, HNPCC5, HSAP; mutS homolog 6 (E. coli); K... 88.6 4e-18
xla:734294 msh6, MGC85188; mutS homolog 6; K08737 DNA mismatch... 87.0 1e-17
cpv:cgd8_370 DNA repair protein ; K08737 DNA mismatch repair p... 85.1 4e-17
bbo:BBOV_I004430 19.m02191; DNA repair protein; K08737 DNA mis... 84.0 1e-16
pfa:PFE0270c DNA repair protein, putative; K08737 DNA mismatch... 82.8 3e-16
ath:AT3G24495 MSH7; MSH7 (MUTS HOMOLOG 7); ATP binding / damag... 80.5 1e-15
ath:AT4G25540 MSH3; MSH3 (Arabidopsis homolog of DNA mismatch ... 78.2 6e-15
cel:H26D21.2 msh-2; MSH (MutS Homolog) family member (msh-2); ... 78.2 7e-15
tpv:TP01_0185 DNA repair protein; K08737 DNA mismatch repair p... 76.3 2e-14
mmu:17686 Msh3, D13Em1, Rep-3, Rep3; mutS homolog 3 (E. coli);... 74.7 6e-14
pfa:PF14_0254 DNA mismatch repair protein Msh2p, putative; K08... 73.9 1e-13
pfa:MAL7P1.206 DNA mismatch repair protein, putative 72.0 4e-13
sce:YCR092C MSH3; Mismatch repair protein, forms dimers with M... 72.0 4e-13
hsa:4437 MSH3, DUP, MGC163306, MGC163308, MRP1; mutS homolog 3... 72.0 4e-13
tgo:TGME49_061000 DNA mismatch repair protein, putative ; K087... 67.8 9e-12
dre:565400 novel protein similar to vertebrate mutS homolog 3 ... 67.0 2e-11
sce:YFL003C MSH4; Msh4p; K08740 DNA mismatch repair protein MSH4 66.6 2e-11
cel:Y47G6A.11 msh-6; MSH (MutS Homolog) family member (msh-6);... 66.2 2e-11
ath:AT4G17380 MSH4; MSH4 (MUTS HOMOLOG 4); ATP binding / damag... 66.2 2e-11
ath:AT3G18524 MSH2; MSH2 (MUTS HOMOLOG 2); ATP binding / damag... 65.5 5e-11
eco:b2733 mutS, ant, ECK2728, fdv, JW2703, plm; methyl-directe... 64.7 7e-11
sce:YOL090W MSH2, PMS5; Msh2p; K08735 DNA mismatch repair prot... 63.5 1e-10
mmu:55993 Msh4, 4930485C04Rik, AV144863, mMsh4; mutS homolog 4... 62.4 4e-10
tpv:TP01_0456 DNA mismatch repair protein MSH2; K08735 DNA mis... 62.0 4e-10
sce:YDL154W MSH5; Msh5p; K08741 DNA mismatch repair protein MSH5 62.0 4e-10
dre:406845 msh2, wu:fc06b02, wu:fc13e09, zgc:55333; mutS homol... 62.0 4e-10
hsa:4438 MSH4; mutS homolog 4 (E. coli); K08740 DNA mismatch r... 62.0 5e-10
ath:AT4G02070 MSH6; MSH6 (MUTS HOMOLOG 6); damaged DNA binding... 61.2 8e-10
cpv:cgd8_3950 MutS like ABC ATpase involved in DNA repair ; K0... 60.8 9e-10
hsa:4436 MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2; mutS homolog... 60.5 1e-09
ath:AT3G20475 MSH5; MSH5 (MUTS-HOMOLOGUE 5); ATP binding / dam... 60.1 2e-09
mmu:17685 Msh2, AI788990; mutS homolog 2 (E. coli); K08735 DNA... 58.5 5e-09
mmu:100046843 DNA mismatch repair protein Msh3-like 57.8 8e-09
hsa:4439 MSH5, DKFZp434C1615, G7, MGC2939, MUTSH5, NG23; mutS ... 57.8 9e-09
mmu:17687 Msh5, G7, Mut5; mutS homolog 5 (E. coli); K08741 DNA... 57.4 1e-08
sce:YHR120W MSH1; DNA-binding protein of the mitochondria invo... 57.0 1e-08
bbo:BBOV_I003810 19.m02277; DNA mismatch repair enzyme; K08735... 56.6 2e-08
dre:559917 mutS homolog 4-like; K08740 DNA mismatch repair pro... 53.9 1e-07
cel:ZK1127.11 him-14; High Incidence of Males (increased X chr... 51.2 8e-07
sce:YDR097C MSH6, PMS3; Msh6p; K08737 DNA mismatch repair prot... 50.8 1e-06
dre:570030 msh5; mutS homolog 5 (E. coli); K08741 DNA mismatch... 48.5 6e-06
cel:F09E8.3 msh-5; MSH (MutS Homolog) family member (msh-5); K... 45.1 6e-05
ath:AT5G54090 DNA mismatch repair MutS family protein 42.7 3e-04
ath:AT3G24320 MSH1; MSH1 (MUTL PROTEIN HOMOLOG 1); ATP binding... 36.2 0.028
tgo:TGME49_018840 mismatch repair protein, putative (EC:2.7.1.... 32.0 0.48
cpv:cgd7_2660 RAD24/Rf-C activator 1 AAA+ ATpase ; K06662 cell... 31.2 0.75
pfa:PF14_0051 DNA mismatch repair protein, putative 31.2 0.82
> tgo:TGME49_090640 DNA mismatch repair protein, putative (EC:3.2.1.3);
K08737 DNA mismatch repair protein MSH6
Length=1607
Score = 106 bits (265), Expect = 2e-23, Method: Composition-based stats.
Identities = 54/97 (55%), Positives = 64/97 (65%), Gaps = 13/97 (13%)
Query 17 MARPKLLPPSEDEEPVLILRNARHPIAEKLTENFVPNDVMLNVPADYSSSSNSSSSSSSS 76
M RP ++ ED P+L L+N RHP+AE L +NFVPNDV LN S
Sbjct 1288 MCRPTIVEAREDAPPILALKNCRHPVAETLMDNFVPNDVYLNCGPHESK----------- 1336
Query 77 KTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
RTLLITGPNMGGKSTLLRQ ALCV++AQ+G +VPA
Sbjct 1337 --RTLLITGPNMGGKSTLLRQAALCVVMAQVGCFVPA 1371
> mmu:17688 Msh6, AU044881, AW550279, GTBP, Gtmbp; mutS homolog
6 (E. coli); K08737 DNA mismatch repair protein MSH6
Length=1358
Score = 94.4 bits (233), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 46/115 (40%), Positives = 67/115 (58%), Gaps = 12/115 (10%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEK--LTENFVPNDVMLN 58
VL LA +++ GP M RP+++ P ED P L + +RHP K ++F+PND+++
Sbjct 1056 VLLCLANYSQGGDGP-MCRPEIVLPGEDTHPFLEFKGSRHPCITKTFFGDDFIPNDILIG 1114
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ + K +L+TGPNMGGKSTL+RQ L ++AQLG YVPA
Sbjct 1115 CEEE---------AEEHGKAYCVLVTGPNMGGKSTLIRQAGLLAVMAQLGCYVPA 1160
> dre:260437 msh6, cb326, wu:fb36c12; mutS homolog 6 (E. coli);
K08737 DNA mismatch repair protein MSH6
Length=1369
Score = 92.8 bits (229), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 49/117 (41%), Positives = 71/117 (60%), Gaps = 9/117 (7%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDE--EPVLILRNARHPIAEK--LTENFVPNDVM 56
VL S+ ++++ G MARP++ P + P L LR +RHP K ++F+PND+
Sbjct 1060 VLLSMCRYSQSADG-SMARPEMALPGDGSYSAPFLDLRGSRHPCVTKTFFGDDFIPNDIF 1118
Query 57 LNVPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ P D + + + + + +L+TGPNMGGKSTL+RQ L VILAQLG YVPA
Sbjct 1119 IGCPGDEEEAQDDTKALAP----CVLVTGPNMGGKSTLMRQCGLVVILAQLGCYVPA 1171
> hsa:2956 MSH6, GTBP, HNPCC5, HSAP; mutS homolog 6 (E. coli);
K08737 DNA mismatch repair protein MSH6
Length=1360
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 47/115 (40%), Positives = 67/115 (58%), Gaps = 13/115 (11%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEK--LTENFVPNDVMLN 58
VL LA +++ GP M RP +L P ED P L L+ +RHP K ++F+PND+++
Sbjct 1059 VLLCLANYSRGGDGP-MCRPVILLP-EDTPPFLELKGSRHPCITKTFFGDDFIPNDILIG 1116
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ + K +L+TGPNMGGKSTL+RQ L ++AQ+G YVPA
Sbjct 1117 CEEE---------EQENGKAYCVLVTGPNMGGKSTLMRQAGLLAVMAQMGCYVPA 1162
> xla:734294 msh6, MGC85188; mutS homolog 6; K08737 DNA mismatch
repair protein MSH6
Length=1340
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 47/115 (40%), Positives = 70/115 (60%), Gaps = 11/115 (9%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEK--LTENFVPNDVMLN 58
VL SL+ +++ GP + RP+++ E+ P L L+ +RHP K ++F+PND+++
Sbjct 1039 VLISLSQYSQGGDGP-VCRPEIVL-QENGSPFLELKGSRHPCITKTFFGDDFIPNDILVG 1096
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ S S + +L+TGPNMGGKSTLLRQ L V++AQLG YVPA
Sbjct 1097 CKEE-------DSDDGSDEAHCVLVTGPNMGGKSTLLRQAGLQVVMAQLGCYVPA 1144
> cpv:cgd8_370 DNA repair protein ; K08737 DNA mismatch repair
protein MSH6
Length=1242
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 51/115 (44%), Positives = 67/115 (58%), Gaps = 17/115 (14%)
Query 1 VLQSLATFA-KTHPGPGMARPKLLPPSE-DEEPVLILRNARHPIAEKLTENFVPNDVMLN 58
VL SLA + T GP +P L E + P+L L+ +RHP+ KL N++PND++LN
Sbjct 939 VLSSLALVSLDTSDGP-FCKPVFLSKEETNGLPMLELKESRHPVVAKLKTNYIPNDILLN 997
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
SS L+TGPNMGGKST+LRQT + VI+AQ+G YVPA
Sbjct 998 --------------GGSSPAPCSLVTGPNMGGKSTILRQTCISVIMAQIGCYVPA 1038
> bbo:BBOV_I004430 19.m02191; DNA repair protein; K08737 DNA mismatch
repair protein MSH6
Length=1313
Score = 84.0 bits (206), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 51/114 (44%), Positives = 66/114 (57%), Gaps = 18/114 (15%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLT-ENFVPNDVMLNV 59
L SLA AK P + R +++ EP +++R+A HPI ++ E FVPNDV L
Sbjct 1023 CLCSLAAVAKNSVLP-LTRAEVIDRGT-AEPFVLIRDAVHPIVSQIDPEGFVPNDVQLG- 1079
Query 60 PADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
DY +L+TGPNMGGKSTLLRQ ALCVI+AQ+GS+VP
Sbjct 1080 HGDYQP--------------LILLTGPNMGGKSTLLRQVALCVIMAQMGSFVPG 1119
> pfa:PFE0270c DNA repair protein, putative; K08737 DNA mismatch
repair protein MSH6
Length=1350
Score = 82.8 bits (203), Expect = 3e-16, Method: Composition-based stats.
Identities = 48/115 (41%), Positives = 62/115 (53%), Gaps = 19/115 (16%)
Query 2 LQSLATFAKTHPGPGMARPKLLP----PSEDEEPVLILRNARHPIAEKLTENFVPNDVML 57
LQS A P + RP L P E++ P LIL + HP+ L NF+ N++ +
Sbjct 1053 LQSFAYVVLNTAFP-LTRPILHPMDSKQGEEKTPFLILEDNIHPVVAMLMPNFISNNIYM 1111
Query 58 NVPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
K TLL+TGPNMGGKSTLLRQTA+ VILAQ+G++VP
Sbjct 1112 --------------GCDKEKQSTLLLTGPNMGGKSTLLRQTAISVILAQIGAFVP 1152
> ath:AT3G24495 MSH7; MSH7 (MUTS HOMOLOG 7); ATP binding / damaged
DNA binding / mismatched DNA binding
Length=1109
Score = 80.5 bits (197), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 53/121 (43%), Positives = 64/121 (52%), Gaps = 18/121 (14%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEE-------PVLILRNARHPIAEKLTENF-VP 52
VL+S A A G MARP + P SE + P+L ++ HP A VP
Sbjct 771 VLRSFAIAASLSAGS-MARPVIFPESEATDQNQKTKGPILKIQGLWHPFAVAADGQLPVP 829
Query 53 NDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
ND++L + SS S R+LL+TGPNMGGKSTLLR T L VI AQLG YVP
Sbjct 830 NDILLG---------EARRSSGSIHPRSLLLTGPNMGGKSTLLRATCLAVIFAQLGCYVP 880
Query 113 A 113
Sbjct 881 C 881
> ath:AT4G25540 MSH3; MSH3 (Arabidopsis homolog of DNA mismatch
repair protein MSH3); damaged DNA binding / mismatched DNA
binding / protein binding; K08736 DNA mismatch repair protein
MSH3
Length=1081
Score = 78.2 bits (191), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 46/114 (40%), Positives = 65/114 (57%), Gaps = 24/114 (21%)
Query 2 LQSLATFAKTHPGPGMARPKLLPPSEDEEPVLI-LRNARHPIAEK-LTENFVPNDVMLNV 59
L SL+T ++ RP+ + +D EPV I +++ RHP+ E L +NFVPND +L+
Sbjct 754 LHSLSTLSRN---KNYVRPEFV---DDCEPVEINIQSGRHPVLETILQDNFVPNDTILHA 807
Query 60 PADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+Y +ITGPNMGGKS +RQ AL I+AQ+GS+VPA
Sbjct 808 EGEYCQ----------------IITGPNMGGKSCYIRQVALISIMAQVGSFVPA 845
> cel:H26D21.2 msh-2; MSH (MutS Homolog) family member (msh-2);
K08735 DNA mismatch repair protein MSH2
Length=849
Score = 78.2 bits (191), Expect = 7e-15, Method: Composition-based stats.
Identities = 48/113 (42%), Positives = 62/113 (54%), Gaps = 23/113 (20%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTEN-FVPNDVMLNV 59
V SL+TFA T G RP LLP L L+ RHP+ E +E F+PNDV+L+
Sbjct 576 VFVSLSTFAATSSGI-YTRPNLLPLGSKR---LELKQCRHPVIEGNSEKPFIPNDVVLD- 630
Query 60 PADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
K R +++TG NMGGKST LR AL ++LAQ+GS+VP
Sbjct 631 -----------------KCRLIILTGANMGGKSTYLRSAALSILLAQIGSFVP 666
> tpv:TP01_0185 DNA repair protein; K08737 DNA mismatch repair
protein MSH6
Length=1160
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 49/110 (44%), Positives = 63/110 (57%), Gaps = 19/110 (17%)
Query 2 LQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTENFVPNDVMLNVPA 61
L SLAT AK P P M RPKL P S++ +L ++++ +PI FVPN V N+
Sbjct 864 LTSLATVAKNSPFP-MCRPKLHPKSQN---ILRVKDSVYPIFSISGNKFVPNSV--NIGE 917
Query 62 DYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYV 111
+ L+ITGPNMGGKSTLLRQ AL VI+ Q+GS+V
Sbjct 918 GFDGP-------------ILIITGPNMGGKSTLLRQIALTVIMGQIGSFV 954
> mmu:17686 Msh3, D13Em1, Rep-3, Rep3; mutS homolog 3 (E. coli);
K08736 DNA mismatch repair protein MSH3
Length=1095
Score = 74.7 bits (182), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 43/115 (37%), Positives = 60/115 (52%), Gaps = 24/115 (20%)
Query 2 LQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKL---TENFVPNDVMLN 58
+ + + AK RP L EE +I++N RHP+ + L + FVPN
Sbjct 789 VDCIFSLAKVAKQGNYCRPTL-----QEEKKIIIKNGRHPMIDVLLGEQDQFVPN----- 838
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
S+S S R ++ITGPNMGGKS+ ++Q AL I+AQ+GSYVPA
Sbjct 839 -----------STSLSQDSERVMIITGPNMGGKSSYIKQVALVTIMAQIGSYVPA 882
> pfa:PF14_0254 DNA mismatch repair protein Msh2p, putative; K08735
DNA mismatch repair protein MSH2
Length=811
Score = 73.9 bits (180), Expect = 1e-13, Method: Composition-based stats.
Identities = 42/114 (36%), Positives = 61/114 (53%), Gaps = 23/114 (20%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKL--TENFVPNDVMLN 58
VL S + P P RP ++ E+ +I+R +RHP+ E NF+PND+ +N
Sbjct 495 VLISFSVVCHNSPFP-YVRPVIVDHGEN----VIMRKSRHPLLELQYNLNNFIPNDIHMN 549
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
+R +++TGPNMGGKST +RQTA+ ILAQ+G +VP
Sbjct 550 ----------------KKNSRLIIVTGPNMGGKSTYIRQTAIICILAQIGMFVP 587
> pfa:MAL7P1.206 DNA mismatch repair protein, putative
Length=873
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 37/92 (40%), Positives = 51/92 (55%), Gaps = 19/92 (20%)
Query 24 PPSEDEEPVLILRNARHPIAEK---LTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRT 80
P E+ +L ++N+RHP+ E L NF+PNDV +N TR
Sbjct 575 PEIEENGHILHMKNSRHPLVESNLLLINNFIPNDVYMN----------------KDITRL 618
Query 81 LLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
+ITGPNMGGKST +RQ AL ++A +G +VP
Sbjct 619 NIITGPNMGGKSTYIRQIALICVMAHIGCFVP 650
> sce:YCR092C MSH3; Mismatch repair protein, forms dimers with
Msh2p that mediate repair of insertion or deletion mutations
and removal of nonhomologous DNA ends, contains a PCNA (Pol30p)
binding motif required for genome stability; K08736 DNA
mismatch repair protein MSH3
Length=1018
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 37/90 (41%), Positives = 53/90 (58%), Gaps = 16/90 (17%)
Query 24 PPSEDEEPVLILRNARHPIAEKLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLI 83
P + + +I +NAR+PI E L ++VPND+M+ S + +I
Sbjct 746 PTFVNGQQAIIAKNARNPIIESLDVHYVPNDIMM----------------SPENGKINII 789
Query 84 TGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
TGPNMGGKS+ +RQ AL I+AQ+GS+VPA
Sbjct 790 TGPNMGGKSSYIRQVALLTIMAQIGSFVPA 819
> hsa:4437 MSH3, DUP, MGC163306, MGC163308, MRP1; mutS homolog
3 (E. coli); K08736 DNA mismatch repair protein MSH3
Length=1137
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 36/92 (39%), Positives = 53/92 (57%), Gaps = 19/92 (20%)
Query 25 PSEDEEPVLILRNARHPIAEKL---TENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTL 81
P+ EE ++++N RHP+ + L + +VPN+ L S R +
Sbjct 849 PTVQEERKIVIKNGRHPVIDVLLGEQDQYVPNNTDL----------------SEDSERVM 892
Query 82 LITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ITGPNMGGKS+ ++Q AL I+AQ+GSYVPA
Sbjct 893 IITGPNMGGKSSYIKQVALITIMAQIGSYVPA 924
> tgo:TGME49_061000 DNA mismatch repair protein, putative ; K08735
DNA mismatch repair protein MSH2
Length=936
Score = 67.8 bits (164), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 39/97 (40%), Positives = 56/97 (57%), Gaps = 22/97 (22%)
Query 19 RPKLLPPSEDEEPVLILRNARHPIAEKL--TENFVPNDVMLNVPADYSSSSNSSSSSSSS 76
RP+++ E + L+L+ +RHP+ E T +F+ NDV L+
Sbjct 574 RPQIV---EGDAGGLVLKASRHPLLEIQPGTSSFIANDVHLD-----------------R 613
Query 77 KTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ R +ITGPNMGGKST +RQ AL V+LAQ+GS+VP
Sbjct 614 ERRLAIITGPNMGGKSTYIRQVALTVLLAQIGSFVPC 650
> dre:565400 novel protein similar to vertebrate mutS homolog
3 (E. coli) (MSH3); K08736 DNA mismatch repair protein MSH3
Length=1083
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 42/115 (36%), Positives = 60/115 (52%), Gaps = 26/115 (22%)
Query 2 LQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKL---TENFVPNDVMLN 58
L SLA AK + RP++L +E+ +++ +HP+ L + +VPND L
Sbjct 789 LFSLAQVAKEN---NYCRPEVL----EEKSQILITAGKHPVITSLMGDQDQYVPNDTHLQ 841
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
R ++ITGPNMGGKS+ +RQ AL I+AQLGS+VPA
Sbjct 842 ----------------GDGKRAMIITGPNMGGKSSYIRQVALVTIMAQLGSFVPA 880
> sce:YFL003C MSH4; Msh4p; K08740 DNA mismatch repair protein
MSH4
Length=878
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/81 (44%), Positives = 51/81 (62%), Gaps = 16/81 (19%)
Query 33 LILRNARHPIAEKLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKS 92
L++R++RHP+ EK+ +NFVPN + SS+ +SSS +ITG NM GKS
Sbjct 598 LLIRDSRHPLLEKVLKNFVPNTI--------SSTKHSSSLQ--------IITGCNMSGKS 641
Query 93 TLLRQTALCVILAQLGSYVPA 113
L+Q AL I+AQ+GS +PA
Sbjct 642 VYLKQVALICIMAQMGSGIPA 662
> cel:Y47G6A.11 msh-6; MSH (MutS Homolog) family member (msh-6);
K08737 DNA mismatch repair protein MSH6
Length=1186
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 46/113 (40%), Positives = 62/113 (54%), Gaps = 12/113 (10%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTENFVPNDVMLNVP 60
VL SLA FAK+ P M P+ + D P LI+ HP + N V
Sbjct 890 VLTSLALFAKSSPFE-MCMPEFDFNATD--PYLIVDKGVHPCLALQSRNEVTQ------- 939
Query 61 ADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+S +S++ +S+ +L+TGPNMGGKSTL+RQTA+ ILA +GS VPA
Sbjct 940 --TTSFIANSTTMGASEAAVMLLTGPNMGGKSTLMRQTAVLAILAHIGSMVPA 990
> ath:AT4G17380 MSH4; MSH4 (MUTS HOMOLOG 4); ATP binding / damaged
DNA binding / mismatched DNA binding; K08740 DNA mismatch
repair protein MSH4
Length=792
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 42/113 (37%), Positives = 55/113 (48%), Gaps = 22/113 (19%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTENFVPNDVMLNVP 60
++ S A T P +RP+L D P L + RHPI E + +FV N + +
Sbjct 491 IVNSFAHTISTKPVDRYSRPEL----TDSGP-LAIDAGRHPILESIHNDFVSNSIFM--- 542
Query 61 ADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
S T L++ GPNM GKST L+Q L VILAQ+G YVPA
Sbjct 543 --------------SEATNMLVVMGPNMSGKSTYLQQVCLVVILAQIGCYVPA 581
> ath:AT3G18524 MSH2; MSH2 (MUTS HOMOLOG 2); ATP binding / damaged
DNA binding / mismatched DNA binding / protein binding;
K08735 DNA mismatch repair protein MSH2
Length=937
Score = 65.5 bits (158), Expect = 5e-11, Method: Composition-based stats.
Identities = 40/113 (35%), Positives = 58/113 (51%), Gaps = 21/113 (18%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLT-ENFVPNDVMLNV 59
VL S A A + P P RP++ + + ++L +RHP E NF+PND L
Sbjct 601 VLLSFADLAASCPTP-YCRPEI---TSSDAGDIVLEGSRHPCVEAQDWVNFIPNDCRL-- 654
Query 60 PADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
K+ ++TGPNMGGKST +RQ + V++AQ+GS+VP
Sbjct 655 --------------MRGKSWFQIVTGPNMGGKSTFIRQVGVIVLMAQVGSFVP 693
> eco:b2733 mutS, ant, ECK2728, fdv, JW2703, plm; methyl-directed
mismatch repair protein; K03555 DNA mismatch repair protein
MutS
Length=853
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 52/90 (57%), Gaps = 18/90 (20%)
Query 25 PSEDEEPVLILRNARHPIAEK-LTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLI 83
P+ ++P + + RHP+ E+ L E F+ N + L S + R L+I
Sbjct 570 PTFIDKPGIRITEGRHPVVEQVLNEPFIANPLNL-----------------SPQRRMLII 612
Query 84 TGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
TGPNMGGKST +RQTAL ++A +GSYVPA
Sbjct 613 TGPNMGGKSTYMRQTALIALMAYIGSYVPA 642
> sce:YOL090W MSH2, PMS5; Msh2p; K08735 DNA mismatch repair protein
MSH2
Length=964
Score = 63.5 bits (153), Expect = 1e-10, Method: Composition-based stats.
Identities = 44/115 (38%), Positives = 61/115 (53%), Gaps = 23/115 (20%)
Query 2 LQSLATFAKTH---PGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTE-NFVPNDVML 57
L +A+FA T P P + RPKL P + LI ++RHP+ E + +F+ NDV L
Sbjct 620 LDVIASFAHTSSYAPIPYI-RPKLHPMDSERRTHLI--SSRHPVLEMQDDISFISNDVTL 676
Query 58 NVPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
S K L+ITGPNMGGKST +RQ + ++AQ+G +VP
Sbjct 677 ----------------ESGKGDFLIITGPNMGGKSTYIRQVGVISLMAQIGCFVP 715
> mmu:55993 Msh4, 4930485C04Rik, AV144863, mMsh4; mutS homolog
4 (E. coli); K08740 DNA mismatch repair protein MSH4
Length=958
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 42/114 (36%), Positives = 56/114 (49%), Gaps = 25/114 (21%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLT-ENFVPNDVMLNV 59
+L L +FA RP+ L ++ HPI EK++ E V N+ +
Sbjct 641 MLDMLLSFAHACTLSDYVRPEF-------TDTLAIKQGWHPILEKISAEKPVANNTYI-- 691
Query 60 PADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ SN L+ITGPNM GKST L+Q ALC I+AQ+GSYVPA
Sbjct 692 ----TEGSN-----------VLIITGPNMSGKSTYLKQIALCQIMAQIGSYVPA 730
> tpv:TP01_0456 DNA mismatch repair protein MSH2; K08735 DNA mismatch
repair protein MSH2
Length=790
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 34/91 (37%), Positives = 50/91 (54%), Gaps = 19/91 (20%)
Query 25 PSEDEEPVLI-LRNARHPIAEKL--TENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTL 81
P+ DE + L ARHP+ E + + +F+PND+ + ++R
Sbjct 524 PTIDETGKTVNLTEARHPLVEYVLSSNSFIPNDLYM----------------ERDRSRVQ 567
Query 82 LITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
+ITGPNMGGKST ++Q L IL Q+GS+VP
Sbjct 568 IITGPNMGGKSTYIKQIGLIAILNQIGSFVP 598
> sce:YDL154W MSH5; Msh5p; K08741 DNA mismatch repair protein
MSH5
Length=901
Score = 62.0 bits (149), Expect = 4e-10, Method: Composition-based stats.
Identities = 35/106 (33%), Positives = 55/106 (51%), Gaps = 9/106 (8%)
Query 8 FAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTENFVPNDVMLNVPADYSSSS 67
FA+ A P+L+ ++E +L + N RH + E +N++PN M++
Sbjct 575 FAQVSAERNYAEPQLV----EDECILEIINGRHALYETFLDNYIPNSTMID-----GGLF 625
Query 68 NSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ S +K R +++TG N GKS L Q L V LAQ+G +VPA
Sbjct 626 SELSWCEQNKGRIIVVTGANASGKSVYLTQNGLIVYLAQIGCFVPA 671
> dre:406845 msh2, wu:fc06b02, wu:fc13e09, zgc:55333; mutS homolog
2 (E. coli); K08735 DNA mismatch repair protein MSH2
Length=936
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/111 (35%), Positives = 53/111 (47%), Gaps = 21/111 (18%)
Query 4 SLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTE-NFVPNDVMLNVPAD 62
S A + P P RPK+L L+L+ ARHP E E F+PNDV
Sbjct 607 SFAVVSHAAPVP-FIRPKILEKGSGR---LVLKAARHPCVEAQDEVAFIPNDVTF----- 657
Query 63 YSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ +ITGP+MGGKST +RQ + V++AQ+G +VP
Sbjct 658 -----------IRGEKMFHIITGPSMGGKSTYIRQVGVIVLMAQIGCFVPC 697
> hsa:4438 MSH4; mutS homolog 4 (E. coli); K08740 DNA mismatch
repair protein MSH4
Length=936
Score = 62.0 bits (149), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 56/114 (49%), Gaps = 25/114 (21%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLT-ENFVPNDVMLNV 59
+L L +FA RP+ L ++ HPI EK++ E + N+ +
Sbjct 619 MLDMLLSFAHACTLSDYVRPEF-------TDTLAIKQGWHPILEKISAEKPIANNTYVTE 671
Query 60 PADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+++ L+ITGPNM GKST L+Q ALC I+AQ+GSYVPA
Sbjct 672 GSNF-----------------LIITGPNMSGKSTYLKQIALCQIMAQIGSYVPA 708
> ath:AT4G02070 MSH6; MSH6 (MUTS HOMOLOG 6); damaged DNA binding;
K08737 DNA mismatch repair protein MSH6
Length=1324
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 45/116 (38%), Positives = 60/116 (51%), Gaps = 19/116 (16%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPI--AEKLTE-NFVPNDVML 57
VL SLA + ++ G RP + + D P L HP+ + L +FVPN+V +
Sbjct 1012 VLISLAFASDSYEGV-RCRPVISGSTSDGVPHLSATGLGHPVLRGDSLGRGSFVPNNVKI 1070
Query 58 NVPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+ K +L+TGPNMGGKSTLLRQ L VILAQ+G+ VPA
Sbjct 1071 G---------------GAEKASFILLTGPNMGGKSTLLRQVCLAVILAQIGADVPA 1111
> cpv:cgd8_3950 MutS like ABC ATpase involved in DNA repair ;
K08735 DNA mismatch repair protein MSH2
Length=848
Score = 60.8 bits (146), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 34/83 (40%), Positives = 44/83 (53%), Gaps = 21/83 (25%)
Query 36 RNARHPIAE-----KLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNMGG 90
+ RHP+ E T FV NDV L+ + S +ITGPNMGG
Sbjct 549 KELRHPLIEAQGTVSTTGQFVANDVELHRHGNLLS----------------IITGPNMGG 592
Query 91 KSTLLRQTALCVILAQLGSYVPA 113
KST +RQ A+C +LAQ+G +VPA
Sbjct 593 KSTYIRQIAICSLLAQIGCFVPA 615
> hsa:4436 MSH2, COCA1, FCC1, HNPCC, HNPCC1, LCFS2; mutS homolog
2, colon cancer, nonpolyposis type 1 (E. coli); K08735 DNA
mismatch repair protein MSH2
Length=934
Score = 60.5 bits (145), Expect = 1e-09, Method: Composition-based stats.
Identities = 40/114 (35%), Positives = 57/114 (50%), Gaps = 22/114 (19%)
Query 2 LQSLATFAKTHPGPGM--ARPKLLPPSEDEEPVLILRNARHPIAEKLTE-NFVPNDVMLN 58
L ++ +FA G + RP +L E + +IL+ +RH E E F+PNDV
Sbjct 602 LDAVVSFAHVSNGAPVPYVRPAIL---EKGQGRIILKASRHACVEVQDEIAFIPNDVYF- 657
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
K +ITGPNMGGKST +RQT + V++AQ+G +VP
Sbjct 658 ---------------EKDKQMFHIITGPNMGGKSTYIRQTGVIVLMAQIGCFVP 696
> ath:AT3G20475 MSH5; MSH5 (MUTS-HOMOLOGUE 5); ATP binding / damaged
DNA binding / mismatched DNA binding; K08741 DNA mismatch
repair protein MSH5
Length=807
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 31/84 (36%), Positives = 43/84 (51%), Gaps = 17/84 (20%)
Query 30 EPVLILRNARHPIAEKLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNMG 89
E +L +RN RH + E + F+PND +N R +ITGPN
Sbjct 531 ESLLDIRNGRHVLQEMAVDTFIPNDTEIN-----------------DNGRIHIITGPNYS 573
Query 90 GKSTLLRQTALCVILAQLGSYVPA 113
GKS ++Q AL V L+ +GS+VPA
Sbjct 574 GKSIYVKQVALIVFLSHIGSFVPA 597
> mmu:17685 Msh2, AI788990; mutS homolog 2 (E. coli); K08735 DNA
mismatch repair protein MSH2
Length=935
Score = 58.5 bits (140), Expect = 5e-09, Method: Composition-based stats.
Identities = 40/110 (36%), Positives = 54/110 (49%), Gaps = 21/110 (19%)
Query 4 SLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTE-NFVPNDVMLNVPAD 62
S A + P P RP +L E + +IL+ +RH E E F+PNDV
Sbjct 607 SFAHVSNAAPVP-YVRPVIL---EKGKGRIILKASRHACVEVQDEVAFIPNDVHF----- 657
Query 63 YSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
K +ITGPNMGGKST +RQT + V++AQ+G +VP
Sbjct 658 -----------EKDKQMFHIITGPNMGGKSTYIRQTGVIVLMAQIGCFVP 696
> mmu:100046843 DNA mismatch repair protein Msh3-like
Length=961
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 24/43 (55%), Positives = 31/43 (72%), Gaps = 0/43 (0%)
Query 71 SSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
S R ++ITGPNMGGKS+ ++Q AL I+AQ+GSYVPA
Sbjct 706 SGQEQDSERVMIITGPNMGGKSSYIKQVALVTIMAQIGSYVPA 748
> hsa:4439 MSH5, DKFZp434C1615, G7, MGC2939, MUTSH5, NG23; mutS
homolog 5 (E. coli); K08741 DNA mismatch repair protein MSH5
Length=834
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 41/115 (35%), Positives = 55/115 (47%), Gaps = 27/115 (23%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILR--NARHPIAEKLTENFVPNDVMLN 58
VL +LA+ A+ + G +RP+ P VL +R N RHP+ E FVPN
Sbjct 531 VLLALASAARDY---GYSRPRYSPQ------VLGVRIQNGRHPLMELCARTFVPN----- 576
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
S+ K R +ITGPN GKS L+Q L +A +GS+VPA
Sbjct 577 -----------STECGGDKGRVKVITGPNSSGKSIYLKQVGLITFMALVGSFVPA 620
> mmu:17687 Msh5, G7, Mut5; mutS homolog 5 (E. coli); K08741 DNA
mismatch repair protein MSH5
Length=833
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 38/113 (33%), Positives = 52/113 (46%), Gaps = 23/113 (20%)
Query 1 VLQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAEKLTENFVPNDVMLNVP 60
VL +LA+ A+ + G +RP P +RN RHP+ E FVPN
Sbjct 530 VLLALASAARDY---GYSRPHYSPCIHGVR----IRNGRHPLMELCARTFVPN------- 575
Query 61 ADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
S+ + R +ITGPN GKS L+Q L +A +GS+VPA
Sbjct 576 ---------STDCGGDQGRVKVITGPNSSGKSIYLKQVGLITFMALVGSFVPA 619
> sce:YHR120W MSH1; DNA-binding protein of the mitochondria involved
in repair of mitochondrial DNA, has ATPase activity and
binds to DNA mismatches; has homology to E. coli MutS; transcription
is induced during meiosis
Length=959
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 37/96 (38%), Positives = 47/96 (48%), Gaps = 23/96 (23%)
Query 22 LLPPSEDEEPVLILRNARHPIAE-----KLTENFVPNDVMLNVPADYSSSSNSSSSSSSS 76
L+ P DE L + N RH + E + E F N+ L +
Sbjct 721 LVCPKVDESNKLEVVNGRHLMVEEGLSARSLETFTANNCEL------------------A 762
Query 77 KTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
K +ITGPNMGGKST LRQ A+ VILAQ+G +VP
Sbjct 763 KDNLWVITGPNMGGKSTFLRQNAIIVILAQIGCFVP 798
> bbo:BBOV_I003810 19.m02277; DNA mismatch repair enzyme; K08735
DNA mismatch repair protein MSH2
Length=791
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 38/113 (33%), Positives = 56/113 (49%), Gaps = 22/113 (19%)
Query 2 LQSLATFAKTHPGPGMARPKLLPPSEDEEPVLILRNARHPIAE--KLTENFVPNDVMLNV 59
L L FA+ RP++ E++E L+ NARHP+ E T FVPN + +
Sbjct 503 LDILVAFAEAAATLQYVRPEI--DLENKEISLV--NARHPLVECGINTRLFVPNSLYM-- 556
Query 60 PADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
+ + + TGPNMGGKST +RQ + V++ Q+GS+VP
Sbjct 557 --------------TRETSLVHITTGPNMGGKSTYIRQVGIIVVMNQIGSFVP 595
> dre:559917 mutS homolog 4-like; K08740 DNA mismatch repair protein
MSH4
Length=931
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 45/81 (55%), Gaps = 16/81 (19%)
Query 33 LILRNARHPIAEKLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKS 92
L ++ RHP+ E+++ S+NS S S+ ++ITGPNM GKS
Sbjct 636 LAIQQGRHPVLERIS-------------GQQPISNNSYISEGSN---FVIITGPNMSGKS 679
Query 93 TLLRQTALCVILAQLGSYVPA 113
T L+Q AL I+AQ+G +VPA
Sbjct 680 TFLKQVALVQIMAQIGCFVPA 700
> cel:ZK1127.11 him-14; High Incidence of Males (increased X chromosome
loss) family member (him-14); K08740 DNA mismatch
repair protein MSH4
Length=842
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 30/84 (35%), Positives = 41/84 (48%), Gaps = 18/84 (21%)
Query 31 PVLILRNARHPIAE-KLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNMG 89
P + RHPI + +E + ND L + R +ITGPNM
Sbjct 550 PSFSISQGRHPILDWDDSEKTITNDTCL-----------------TRDRRFGIITGPNMA 592
Query 90 GKSTLLRQTALCVILAQLGSYVPA 113
GKST L+QTA I+AQ+G ++PA
Sbjct 593 GKSTYLKQTAQLAIMAQIGCFIPA 616
> sce:YDR097C MSH6, PMS3; Msh6p; K08737 DNA mismatch repair protein
MSH6
Length=1242
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 41/85 (48%), Gaps = 19/85 (22%)
Query 32 VLILRNARHP---IAEKLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNM 88
L ++ RHP + ++F+PND+ L + R L+TG N
Sbjct 942 FLKFKSLRHPCFNLGATTAKDFIPNDIEL----------------GKEQPRLGLLTGANA 985
Query 89 GGKSTLLRQTALCVILAQLGSYVPA 113
GKST+LR + VI+AQ+G YVP
Sbjct 986 AGKSTILRMACIAVIMAQMGCYVPC 1010
> dre:570030 msh5; mutS homolog 5 (E. coli); K08741 DNA mismatch
repair protein MSH5
Length=793
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 41/84 (48%), Gaps = 16/84 (19%)
Query 30 EPVLILRNARHPIAEKLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNMG 89
E L L +RHP+ E T FV N + SS ++ + +ITGPN
Sbjct 512 EHRLALIESRHPLLELCTSVFVSNTYI----------------SSETEGKVKVITGPNSS 555
Query 90 GKSTLLRQTALCVILAQLGSYVPA 113
GKS L+Q L V +A +GS VPA
Sbjct 556 GKSIFLKQVGLIVFMALIGSNVPA 579
> cel:F09E8.3 msh-5; MSH (MutS Homolog) family member (msh-5);
K08741 DNA mismatch repair protein MSH5
Length=1369
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 42/86 (48%), Gaps = 17/86 (19%)
Query 29 EEPVLILRNARHPIAEKLTE-NFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPN 87
+EPV+ HPI+ + + +FVPN V SS + +ITGPN
Sbjct 598 DEPVIEAVELYHPISVLVVKKSFVPNQV----------------SSGRDGIKASIITGPN 641
Query 88 MGGKSTLLRQTALCVILAQLGSYVPA 113
GKS ++ + V L+ +GS+VPA
Sbjct 642 ACGKSVYMKSIGIMVFLSHIGSFVPA 667
> ath:AT5G54090 DNA mismatch repair MutS family protein
Length=795
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 9/55 (16%)
Query 59 VPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
+PAD+ S TR L+ITGPN GGK+ L+ L ++A+ G YV A
Sbjct 423 IPADFQISKG---------TRVLVITGPNTGGKTICLKSVGLAAMMAKSGLYVLA 468
> ath:AT3G24320 MSH1; MSH1 (MUTL PROTEIN HOMOLOG 1); ATP binding
/ catalytic/ damaged DNA binding / mismatched DNA binding
/ nuclease
Length=1118
Score = 36.2 bits (82), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 60 PADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVPA 113
P + SS ++ ++ L+TGPN GGKS+LLR +L G VPA
Sbjct 743 PYWFDVSSGTAVHNTVDMQSLFLLTGPNGGGKSSLLRSICAAALLGISGLMVPA 796
> tgo:TGME49_018840 mismatch repair protein, putative (EC:2.7.1.127)
Length=1944
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 44/99 (44%), Gaps = 25/99 (25%)
Query 25 PSEDEEPVLILRNARHPIAEKLTENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRT---- 80
P++DE +R P++E++ ++ ++S + +SSS SK R
Sbjct 1258 PTDDEFANANRTQSRDPLSEQVQKD--------------TASLLAHASSSLSKEREVVSY 1303
Query 81 -------LLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
++TG NM GKST R V+LA + VP
Sbjct 1304 SFDMHGLFVLTGENMAGKSTFCRSVLALVVLANAATTVP 1342
> cpv:cgd7_2660 RAD24/Rf-C activator 1 AAA+ ATpase ; K06662 cell
cycle checkpoint protein
Length=701
Score = 31.2 bits (69), Expect = 0.75, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 48 ENFVPNDVMLNVPADYSSSSNSSSSSSSSKTRTLLITGPNMGGKSTLLR 96
E V ++ V + +S S++S + SK L+I GPN GK+T+LR
Sbjct 15 ELLVNKHKIIEVKSFLENSFISANSINGSKKNILIIFGPNGAGKTTILR 63
> pfa:PF14_0051 DNA mismatch repair protein, putative
Length=1515
Score = 31.2 bits (69), Expect = 0.82, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 77 KTRTLLITGPNMGGKSTLLRQTALCVILAQLGSYVP 112
K +L+TG NM GK+TL + L+ LG Y P
Sbjct 1048 KKNFILLTGKNMSGKTTLSFTILSILFLSNLGMYAP 1083
Lambda K H
0.311 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2052546600
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40