bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2226_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
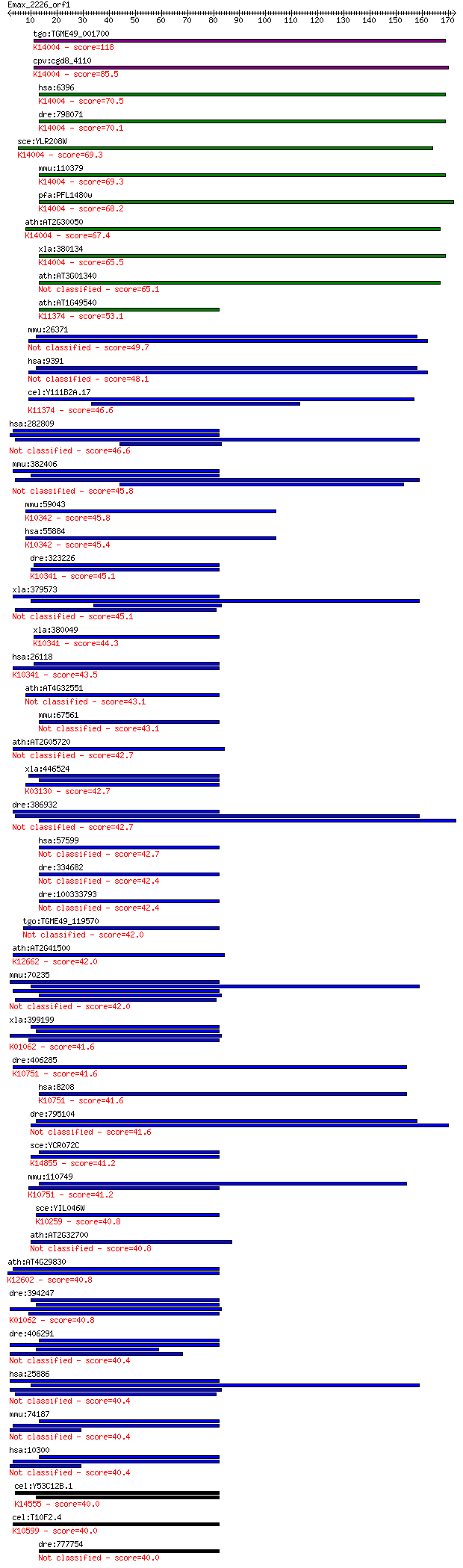
tgo:TGME49_001700 sec 13, putative ; K14004 protein transport ... 118 9e-27
cpv:cgd8_4110 hypothetical protein ; K14004 protein transport ... 85.5 7e-17
hsa:6396 SEC13, D3S1231E, SEC13L1, SEC13R, npp-20; SEC13 homol... 70.5 2e-12
dre:798071 sec13, MGC174049, MGC63980, cb271, sb:cb271, sec13l... 70.1 3e-12
sce:YLR208W SEC13, ANU3; Component of both the Nup84 nuclear p... 69.3 6e-12
mmu:110379 Sec13, 1110003H02Rik, Sec13l1, Sec13r; SEC13 homolo... 69.3 6e-12
pfa:PFL1480w protein transport protein Sec13, putative; K14004... 68.2 1e-11
ath:AT2G30050 transducin family protein / WD-40 repeat family ... 67.4 2e-11
xla:380134 sec13, MGC52900, sec13l1; SEC13 homolog; K14004 pro... 65.5 9e-11
ath:AT3G01340 protein transport protein SEC13 family protein /... 65.1 1e-10
ath:AT1G49540 nucleotide binding; K11374 elongator complex pro... 53.1 4e-07
mmu:26371 Ciao1, AW210570, Wdr39; cytosolic iron-sulfur protei... 49.7 5e-06
hsa:9391 CIAO1, CIA1, WDR39; cytosolic iron-sulfur protein ass... 48.1 2e-05
cel:Y111B2A.17 elpc-2; ELongator complex Protein Component fam... 46.6 4e-05
hsa:282809 POC1B, FLJ14923, FLJ41111, PIX1, TUWD12, WDR51B; PO... 46.6 4e-05
mmu:382406 Poc1b, 4933430F16Rik, Wdr51b; POC1 centriolar prote... 45.8 7e-05
mmu:59043 Wsb2, AA673511, Swip2, WSB-2; WD repeat and SOCS box... 45.8 7e-05
hsa:55884 WSB2, MGC10210, SBA2; WD repeat and SOCS box contain... 45.4 9e-05
dre:323226 wsb1, wu:fb92g11, zgc:63905; WD repeat and SOCS box... 45.1 1e-04
xla:379573 poc1a, MGC69111, pix2, wdr51a; POC1 centriolar prot... 45.1 1e-04
xla:380049 wsb1, MGC53424; WD repeat and SOCS box containing 1... 44.3 2e-04
hsa:26118 WSB1, SWIP1, WSB-1; WD repeat and SOCS box containin... 43.5 3e-04
ath:AT4G32551 LUG; LUG (LEUNIG); protein binding / protein het... 43.1 4e-04
mmu:67561 Wdr48, 8430408H12Rik, mKIAA1449; WD repeat domain 48 43.1
ath:AT2G05720 transducin family protein / WD-40 repeat family ... 42.7 5e-04
xla:446524 taf5l, MGC80243; TAF5-like RNA polymerase II, p300/... 42.7 5e-04
dre:386932 poc1b, TUWD12, fl36w17, wdr51b, wu:fl36w17, zgc:635... 42.7 6e-04
hsa:57599 WDR48, DKFZp686G1794, KIAA1449, P80, UAF1; WD repeat... 42.7 7e-04
dre:334682 wdr48, fk50c12, wd48, wu:fd19a04, wu:fk50c12, zgc:6... 42.4 7e-04
dre:100333793 WD repeat-containing protein 48-like 42.4 8e-04
tgo:TGME49_119570 WD-40 repeat protein, putative (EC:2.7.11.7) 42.0 0.001
ath:AT2G41500 EMB2776; nucleotide binding; K12662 U4/U6 small ... 42.0 0.001
mmu:70235 Poc1a, 2510040D07Rik, Wdr51a; POC1 centriolar protei... 42.0 0.001
xla:399199 pafah1b1, lis2, mdcr, mds, pafah, pafah1b1-b; plate... 41.6 0.001
dre:406285 chaf1b, wu:fd07c09, wu:fe36g06, wu:fv38g09, zgc:560... 41.6 0.001
hsa:8208 CHAF1B, CAF-1, CAF-IP60, CAF1, CAF1A, CAF1P60, MPHOSP... 41.6 0.001
dre:795104 ciao1, MGC55911, MGC77394, wdr39, zgc:55911, zgc:77... 41.6 0.001
sce:YCR072C RSA4; Rsa4p; K14855 ribosome assembly protein 4 41.2
mmu:110749 Chaf1b, 2600017H24Rik, C76145, CAF-I_p60, CAF-Ip60,... 41.2 0.002
sce:YIL046W MET30, ZRG11; F-box protein containing five copies... 40.8 0.002
ath:AT2G32700 LUH; WD-40 repeat family protein 40.8 0.002
ath:AT4G29830 VIP3; VIP3 (vernalization independence 3); nucle... 40.8 0.002
dre:394247 pafah1b1b, Lis1a; platelet-activating factor acetyl... 40.8 0.003
dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin ... 40.4 0.003
hsa:25886 POC1A, DKFZp434C245, MGC131902, PIX2, WDR51A; POC1 c... 40.4 0.003
mmu:74187 Katnb1, 2410003J24Rik, KAT; katanin p80 (WD40-contai... 40.4 0.003
hsa:10300 KATNB1, KAT; katanin p80 (WD repeat containing) subu... 40.4 0.003
cel:Y53C12B.1 hypothetical protein; K14555 U3 small nucleolar ... 40.0 0.003
cel:T10F2.4 hypothetical protein; K10599 pre-mRNA-processing f... 40.0 0.003
dre:777754 zgc:153492 40.0 0.003
> tgo:TGME49_001700 sec 13, putative ; K14004 protein transport
protein SEC13
Length=654
Score = 118 bits (296), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 76/160 (47%), Positives = 98/160 (61%), Gaps = 21/160 (13%)
Query 11 LLLASGGADNRVRLWECDA-SKQWTELPALA-GEHHTDWVRDVSFRPQGASSFVVPGALL 68
L+LA+GG D++VR+W D S++W +L L + HTDWVRDV+F+P ASS ++ + L
Sbjct 210 LMLATGGCDSQVRIWGLDPNSQEWQQLHQLTDADPHTDWVRDVAFQPASASSLLLSSSRL 269
Query 69 LASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTLVLDA 128
LASCSED TVKLW+GE NPS + Y W LLQTL L A
Sbjct 270 LASCSEDGTVKLWVGE-------------------ASTPSANPSATSYTWSLLQTLRLHA 310
Query 129 PAWRVTWSSSGALLAVACGDSAVLLLRENMHGTWELVTDL 168
P WRV+WS SG +L+VACG+ V L RE + G WE V+ L
Sbjct 311 PVWRVSWSVSGTILSVACGEKDVCLFRETVAGHWEKVSRL 350
> cpv:cgd8_4110 hypothetical protein ; K14004 protein transport
protein SEC13
Length=601
Score = 85.5 bits (210), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 51/160 (31%), Positives = 79/160 (49%), Gaps = 34/160 (21%)
Query 11 LLLASGGADNRVRLWECDA-SKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
+ L SGG DN++R+W+ D +K+ +E+ H++WVRDV++RP V A +
Sbjct 178 IRLVSGGCDNQIRIWKQDPQTKELSEMNQTLDVAHSEWVRDVAWRPS-----VDLLAETI 232
Query 70 ASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTLVLDAP 129
ASC +DK V +W + Q W Q L + P
Sbjct 233 ASCGDDKIVVIWTQDADGQ----------------------------GWHSSQVLNFNEP 264
Query 130 AWRVTWSSSGALLAVACGDSAVLLLRENMHGTWELVTDLA 169
WRV+WS +G +LA + G+ V L REN G WE++T+++
Sbjct 265 VWRVSWSVTGTVLAASSGEDVVTLFRENSEGKWEVLTNIS 304
> hsa:6396 SEC13, D3S1231E, SEC13L1, SEC13R, npp-20; SEC13 homolog
(S. cerevisiae); K14004 protein transport protein SEC13
Length=308
Score = 70.5 bits (171), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 51/156 (32%), Positives = 77/156 (49%), Gaps = 34/156 (21%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
ASGG DN ++LW+ + QW E L E H+DWVRDV++ P S +P + +ASC
Sbjct 168 FASGGCDNLIKLWKEEEDGQWKEEQKL--EAHSDWVRDVAWAP----SIGLPTS-TIASC 220
Query 73 SEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTLVLDAPAWR 132
S+D V +W + D N SP +LL + W
Sbjct 221 SQDGRVFIWTCD--------------------DASSNTWSP-----KLLHK--FNDVVWH 253
Query 133 VTWSSSGALLAVACGDSAVLLLRENMHGTWELVTDL 168
V+WS + +LAV+ GD+ V L +E++ G W ++D+
Sbjct 254 VSWSITANILAVSGGDNKVTLWKESVDGQWVCISDV 289
> dre:798071 sec13, MGC174049, MGC63980, cb271, sb:cb271, sec13l1,
wu:fa08f09, zgc:63980; SEC13 homolog (S. cerevisiae); K14004
protein transport protein SEC13
Length=320
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 49/156 (31%), Positives = 77/156 (49%), Gaps = 35/156 (22%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
SGG DN V+LW+ + QW E L E H+DWVRDV + P S +P + + ASC
Sbjct 182 FVSGGCDNLVKLWK-EEDGQWKEDQKL--EAHSDWVRDVGWAP----SIGLPTSTI-ASC 233
Query 73 SEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTLVLDAPAWR 132
S+D V +W ++P+ + + +LL + W
Sbjct 234 SQDGRVFIWT-------------------------CDDPAGNTWTAKLLHKF--NDVVWH 266
Query 133 VTWSSSGALLAVACGDSAVLLLRENMHGTWELVTDL 168
V+WS +G +LAV+ GD+ V L +E++ G W ++D+
Sbjct 267 VSWSITGNILAVSGGDNKVTLWKESVDGQWACISDV 302
> sce:YLR208W SEC13, ANU3; Component of both the Nup84 nuclear
pore sub-complex and the Sec13p-Sec31p complex of the COPII
vesicle coat, required for vesicle formation in ER to Golgi
transport and nuclear pore complex organization; K14004 protein
transport protein SEC13
Length=297
Score = 69.3 bits (168), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 48/159 (30%), Positives = 78/159 (49%), Gaps = 33/159 (20%)
Query 5 DGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVP 64
+G +GGADN V++W+ ++ Q T + E H+DWVRDV++ P V
Sbjct 165 NGTKESRKFVTGGADNLVKIWKYNSDAQ-TYVLESTLEGHSDWVRDVAWSP------TVL 217
Query 65 GALLLASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTL 124
LAS S+D+T +W + +Q ++ L++++ D+L
Sbjct 218 LRSYLASVSQDRTCIIWTQD---NEQGPWKKTLLKEEKFPDVL----------------- 257
Query 125 VLDAPAWRVTWSSSGALLAVACGDSAVLLLRENMHGTWE 163
WR +WS SG +LA++ GD+ V L +EN+ G WE
Sbjct 258 ------WRASWSLSGNVLALSGGDNKVTLWKENLEGKWE 290
> mmu:110379 Sec13, 1110003H02Rik, Sec13l1, Sec13r; SEC13 homolog
(S. cerevisiae); K14004 protein transport protein SEC13
Length=322
Score = 69.3 bits (168), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 48/156 (30%), Positives = 76/156 (48%), Gaps = 34/156 (21%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
ASGG DN ++LW + QW E L E H+DWVRDV++ P S +P + +ASC
Sbjct 182 FASGGCDNLIKLWREEEDGQWKEEQKL--EAHSDWVRDVAWAP----SIGLPTS-TIASC 234
Query 73 SEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTLVLDAPAWR 132
S+D V +W ++ S + + +LL + W
Sbjct 235 SQDGRVFIWT-------------------------CDDASGNMWSPKLLHK--FNDVVWH 267
Query 133 VTWSSSGALLAVACGDSAVLLLRENMHGTWELVTDL 168
V+WS + +LAV+ GD+ V L +E++ G W ++D+
Sbjct 268 VSWSITANILAVSGGDNKVTLWKESVDGQWVCISDV 303
> pfa:PFL1480w protein transport protein Sec13, putative; K14004
protein transport protein SEC13
Length=822
Score = 68.2 bits (165), Expect = 1e-11, Method: Composition-based stats.
Identities = 45/160 (28%), Positives = 79/160 (49%), Gaps = 34/160 (21%)
Query 13 LASGGADNRVRLWECDA-SKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
L SGG DN+V +W D +K++ ++ + + H ++DV+++P +S A ++AS
Sbjct 210 LVSGGYDNQVIIWMFDNNTKEFQKIYQMNDKPHKSSIKDVAWKPNLDNS-----ANIIAS 264
Query 72 CSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTLVLDAPAW 131
CS++K V LWI +D+ N W+ Q + +
Sbjct 265 CSDEKIVILWI---------------------EDVTNN-------VWKNGQIIKVKYNIH 296
Query 132 RVTWSSSGALLAVACGDSAVLLLRENMHGTWELVTDLAEK 171
+++WS +G +LA+AC D L +EN G WE + +L E+
Sbjct 297 KISWSPNGTILAIACSDDNAYLYKENAEGIWEEMCNLTEE 336
> ath:AT2G30050 transducin family protein / WD-40 repeat family
protein; K14004 protein transport protein SEC13
Length=302
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 55/166 (33%), Positives = 86/166 (51%), Gaps = 43/166 (25%)
Query 8 TSGLL-----LASGGADNRVRLWE-CDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
+SGLL LASGG DN V++W+ + S + PAL + HTDWVRDV++ P +
Sbjct 169 SSGLLDPVYKLASGGCDNTVKVWKLANGSWKMDCFPAL--QKHTDWVRDVAWAP----NL 222
Query 62 VVPGALLLASCSEDKTVKLW-IGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRL 120
+P + +AS S+D V +W +G ++ +Q + + +D +
Sbjct 223 GLPKS-TIASGSQDGKVIIWTVG---------KEGEQWEGKVLKDFM------------- 259
Query 121 LQTLVLDAPAWRVTWSSSGALLAVACGDSAVLLLRENMHGTWELVT 166
P WRV+WS +G LLAV+ G++ V + +E + G WE VT
Sbjct 260 -------TPVWRVSWSLTGNLLAVSDGNNNVTVWKEAVDGEWEQVT 298
> xla:380134 sec13, MGC52900, sec13l1; SEC13 homolog; K14004 protein
transport protein SEC13
Length=320
Score = 65.5 bits (158), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 49/156 (31%), Positives = 75/156 (48%), Gaps = 35/156 (22%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
SGG DN V++W + QW E L E H+DWVRDV++ P S +P + +ASC
Sbjct 182 FVSGGCDNLVKIWR-EEDGQWKEDQKL--EAHSDWVRDVAWAP----SIGLPTS-TIASC 233
Query 73 SEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTLVLDAPAWR 132
S+D V +W + D N +P +LL + W
Sbjct 234 SQDGRVYIWTSD--------------------DAATNCWTP-----KLLHK--FNDVVWH 266
Query 133 VTWSSSGALLAVACGDSAVLLLRENMHGTWELVTDL 168
V+WS + +LAV+ GD+ V L +E++ G W ++D+
Sbjct 267 VSWSITANILAVSGGDNKVTLWKESVDGQWACISDV 302
> ath:AT3G01340 protein transport protein SEC13 family protein
/ WD-40 repeat family protein
Length=302
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 52/156 (33%), Positives = 76/156 (48%), Gaps = 38/156 (24%)
Query 13 LASGGADNRVRLWE-CDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
LASGG D+ V++W+ + S + PAL HTDWVRDV++ P + +P + +AS
Sbjct 179 LASGGCDSTVKVWKFSNGSWKMDCFPAL--NKHTDWVRDVAWAP----NLGLPKS-TIAS 231
Query 72 CSEDKTVKLW-IGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTLVLDAPA 130
SED V +W IG+ +Q W P
Sbjct 232 GSEDGKVIIWTIGKEGEQ-----------------------------WEGTVLKDFKTPV 262
Query 131 WRVTWSSSGALLAVACGDSAVLLLRENMHGTWELVT 166
WRV+WS +G LLAV+ G++ V + +E++ G WE VT
Sbjct 263 WRVSWSLTGNLLAVSDGNNNVTVWKESVDGEWEQVT 298
> ath:AT1G49540 nucleotide binding; K11374 elongator complex protein
2
Length=838
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 44/69 (63%), Gaps = 2/69 (2%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
LA GG DN+++L+ + + ++T + L G HTDW+R + F ++ +P +++L S
Sbjct 191 LALGGLDNKIKLYSGERTGKFTSVCELKG--HTDWIRSLDFSLPLHTTEEIPNSIMLVSS 248
Query 73 SEDKTVKLW 81
S+DK +++W
Sbjct 249 SQDKVIRIW 257
> mmu:26371 Ciao1, AW210570, Wdr39; cytosolic iron-sulfur protein
assembly 1 homolog (S. cerevisiae)
Length=339
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 41/148 (27%), Positives = 62/148 (41%), Gaps = 31/148 (20%)
Query 12 LLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
LLAS D+ V+L++ + W L G T W ++F P G LAS
Sbjct 164 LLASASYDDTVKLYQ-EEGDDWVCCATLEGHESTVW--SIAFDPSGQR---------LAS 211
Query 72 CSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTL--VLDAP 129
CS+D+TV++W +Q +Q + + PS W+ + TL
Sbjct 212 CSDDRTVRIW--------------RQYLPGNEQGVACSGSDPS---WKCICTLSGFHTRT 254
Query 130 AWRVTWSSSGALLAVACGDSAVLLLREN 157
+ V W LA ACGD A+ + E+
Sbjct 255 IYDVAWCQLTGALATACGDDAIRVFEED 282
Score = 36.6 bits (83), Expect = 0.042, Method: Compositional matrix adjust.
Identities = 48/204 (23%), Positives = 76/204 (37%), Gaps = 53/204 (25%)
Query 9 SGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQG----ASSFV-- 62
SG LLAS G D ++R+W + W ++ E H VR V++ P G ++SF
Sbjct 27 SGTLLASCGGDRKIRIWGTEG-DSWI-CKSVLSEGHQRTVRKVAWSPCGNYLASASFDAT 84
Query 63 -----------------------------VPGALLLASCSEDKTVKLW-IGEPPQQQ--- 89
P LLA+CS DK+V +W + E + +
Sbjct 85 TCIWKKNQDDFECVTTLEGHENEVKSVAWAPSGNLLATCSRDKSVWVWEVDEEDEYECVS 144
Query 90 ---QHQQQQQQLQQQQQQDILMNNPSPSQYK--------WRLLQTLV-LDAPAWRVTWSS 137
H Q + + Q++L + K W TL ++ W + +
Sbjct 145 VLSSHTQDVKHVVWHPSQELLASASYDDTVKLYQEEGDDWVCCATLEGHESTVWSIAFDP 204
Query 138 SGALLAVACGDSAVLLLRENMHGT 161
SG LA D V + R+ + G
Sbjct 205 SGQRLASCSDDRTVRIWRQYLPGN 228
> hsa:9391 CIAO1, CIA1, WDR39; cytosolic iron-sulfur protein assembly
1
Length=339
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 40/148 (27%), Positives = 62/148 (41%), Gaps = 31/148 (20%)
Query 12 LLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
LLAS D+ V+L+ + W L G T W ++F P G LAS
Sbjct 164 LLASASYDDTVKLYR-EEEDDWVCCATLEGHESTVW--SLAFDPSGQR---------LAS 211
Query 72 CSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTL--VLDAP 129
CS+D+TV++W +Q +Q + + PS W+ + TL
Sbjct 212 CSDDRTVRIW--------------RQYLPGNEQGVACSGSDPS---WKCICTLSGFHSRT 254
Query 130 AWRVTWSSSGALLAVACGDSAVLLLREN 157
+ + W LA ACGD A+ + +E+
Sbjct 255 IYDIAWCQLTGALATACGDDAIRVFQED 282
Score = 35.8 bits (81), Expect = 0.078, Method: Compositional matrix adjust.
Identities = 48/204 (23%), Positives = 76/204 (37%), Gaps = 53/204 (25%)
Query 9 SGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQG----ASSFV-- 62
+G LLAS G D R+R+W + W ++ E H VR V++ P G ++SF
Sbjct 27 AGTLLASCGGDRRIRIWGTEG-DSWI-CKSVLSEGHQRTVRKVAWSPCGNYLASASFDAT 84
Query 63 -----------------------------VPGALLLASCSEDKTVKLW-IGEPPQQQ--- 89
P LLA+CS DK+V +W + E + +
Sbjct 85 TCIWKKNQDDFECVTTLEGHENEVKSVAWAPSGNLLATCSRDKSVWVWEVDEEDEYECVS 144
Query 90 ---QHQQQQQQLQQQQQQDILMNNPSPSQYK--------WRLLQTLV-LDAPAWRVTWSS 137
H Q + + Q++L + K W TL ++ W + +
Sbjct 145 VLNSHTQDVKHVVWHPSQELLASASYDDTVKLYREEEDDWVCCATLEGHESTVWSLAFDP 204
Query 138 SGALLAVACGDSAVLLLRENMHGT 161
SG LA D V + R+ + G
Sbjct 205 SGQRLASCSDDRTVRIWRQYLPGN 228
> cel:Y111B2A.17 elpc-2; ELongator complex Protein Component family
member (elpc-2); K11374 elongator complex protein 2
Length=778
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 39/154 (25%), Positives = 68/154 (44%), Gaps = 15/154 (9%)
Query 9 SGLLLASGGADNRVRLW-ECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGAL 67
+ +LLA G + V L+ E K ++ L ++AG HTDW+ ++F P L
Sbjct 172 NSVLLAVGTSKRFVELYGESADKKSFSRLISVAG--HTDWIHSIAFNDN-------PDHL 222
Query 68 LLASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDIL--MNNPSPSQYKWRLLQTLV 125
L+AS +D V+LW EP ++ + ++ ++ N S + +R V
Sbjct 223 LVASAGQDTYVRLWAIEPETDEKSENIREDSSTTPPDELTSSANLFSINYTPYRCSSHAV 282
Query 126 L---DAPAWRVTWSSSGALLAVACGDSAVLLLRE 156
+ D WS+ G +L A D ++ +E
Sbjct 283 MQGHDDWVHSTVWSNDGRVLLTASSDKTCIIWKE 316
Score = 28.9 bits (63), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 34/82 (41%), Gaps = 11/82 (13%)
Query 33 WTELPALAGEHHTDWVRDVSFR--PQGASSFVVPGALLLASCSEDKTVKLWIGEPPQQQQ 90
WT LP G H VRDV + G S F L S +D+T +++ QQ
Sbjct 383 WTALPMTGG--HVGEVRDVDWHRSDDGDSGF-------LMSVGQDQTTRVFAKNGRQQSY 433
Query 91 HQQQQQQLQQQQQQDILMNNPS 112
+ + Q+ Q + NPS
Sbjct 434 VEIARPQVHGHDMQCLSFVNPS 455
> hsa:282809 POC1B, FLJ14923, FLJ41111, PIX1, TUWD12, WDR51B;
POC1 centriolar protein homolog B (Chlamydomonas)
Length=436
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 35/79 (44%), Gaps = 14/79 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
SVD G LA+ D +++W + L HT WVR F P G
Sbjct 65 SVDFSADGQFLATASEDKSIKVWSMYRQRFLYSL-----YRHTHWVRCAKFSPDGR---- 115
Query 63 VPGALLLASCSEDKTVKLW 81
L+ SCSEDKT+K+W
Sbjct 116 -----LIVSCSEDKTIKIW 129
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 38/80 (47%), Gaps = 14/80 (17%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
+SV G LLAS D VRLW D +++E A HT VR V F G
Sbjct 22 TSVQFSPHGNLLASASRDRTVRLWIPDKRGKFSEFKA-----HTAPVRSVDFSADGQ--- 73
Query 62 VVPGALLLASCSEDKTVKLW 81
LA+ SEDK++K+W
Sbjct 74 ------FLATASEDKSIKVW 87
Score = 37.4 bits (85), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 40/156 (25%), Positives = 58/156 (37%), Gaps = 47/156 (30%)
Query 4 VDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVV 63
VD SG +AS G+D V++W+ +K H+ V +SF P G
Sbjct 150 VDFNPSGTCIASAGSDQTVKVWDVRVNKLLQHYQV-----HSGGVNCISFHPSGN----- 199
Query 64 PGALLLASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQT 123
L + S D T+K+ D+L + RL+ T
Sbjct 200 ----YLITASSDGTLKIL-----------------------DLL---------EGRLIYT 223
Query 124 LV-LDAPAWRVTWSSSGALLAVACGDSAVLLLRENM 158
L P + V++S G L A D+ VLL R N
Sbjct 224 LQGHTGPVFTVSFSKGGELFASGGADTQVLLWRTNF 259
Score = 32.7 bits (73), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 20/39 (51%), Gaps = 9/39 (23%)
Query 44 HTDWVRDVSFRPQGASSFVVPGALLLASCSEDKTVKLWI 82
H D V V F P G LLAS S D+TV+LWI
Sbjct 17 HKDVVTSVQFSPHGN---------LLASASRDRTVRLWI 46
> mmu:382406 Poc1b, 4933430F16Rik, Wdr51b; POC1 centriolar protein
homolog B (Chlamydomonas)
Length=476
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 35/79 (44%), Gaps = 14/79 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
SVD G LL + D +++W + L HT WVR F P G
Sbjct 107 SVDFSADGQLLVTASEDKSIKVWSMFRQRFLYSL-----YRHTHWVRCAKFSPDGR---- 157
Query 63 VPGALLLASCSEDKTVKLW 81
L+ SCSEDKT+K+W
Sbjct 158 -----LIVSCSEDKTIKIW 171
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 34/72 (47%), Gaps = 14/72 (19%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
G LLAS D VRLW D + +E A HT VR V F G LL
Sbjct 72 GNLLASASRDRTVRLWVLDRKGKSSEFKA-----HTAPVRSVDFSADGQ---------LL 117
Query 70 ASCSEDKTVKLW 81
+ SEDK++K+W
Sbjct 118 VTASEDKSIKVW 129
Score = 34.7 bits (78), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 39/156 (25%), Positives = 60/156 (38%), Gaps = 47/156 (30%)
Query 4 VDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVV 63
VD +G +AS G+D+ V++W+ +K H+ V +SF P G S
Sbjct 192 VDFNPNGTCIASAGSDHAVKIWDIRMNKLLQHYQV-----HSCGVNCLSFHPLGNS---- 242
Query 64 PGALLLASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQT 123
L + S D TVK+ D++ + RL+ T
Sbjct 243 -----LVTASSDGTVKML-----------------------DLI---------EGRLIYT 265
Query 124 LV-LDAPAWRVTWSSSGALLAVACGDSAVLLLRENM 158
L P + V++S G LL D+ VL+ R N
Sbjct 266 LQGHTGPVFTVSFSKDGELLTSGGADAQVLIWRTNF 301
Score = 32.3 bits (72), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 46/119 (38%), Gaps = 19/119 (15%)
Query 44 HTDWVRDVSFRPQGASSFVVPGALLLASCSEDKTVKLWI----GEPPQQQQHQQQQQQLQ 99
H D V + F PQG LLAS S D+TV+LW+ G+ + + H + +
Sbjct 59 HKDVVTSLQFSPQGN---------LLASASRDRTVRLWVLDRKGKSSEFKAHTAPVRSVD 109
Query 100 QQQQQDILMNNPSP------SQYKWRLLQTLVLDAPAWRVTWSSSGALLAVACGDSAVL 152
+L+ S ++ R L +L R S L V+C + +
Sbjct 110 FSADGQLLVTASEDKSIKVWSMFRQRFLYSLYRHTHWVRCAKFSPDGRLIVSCSEDKTI 168
> mmu:59043 Wsb2, AA673511, Swip2, WSB-2; WD repeat and SOCS box-containing
2; K10342 WD repeat and SOCS box-containing protein
2
Length=404
Score = 45.8 bits (107), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 47/96 (48%), Gaps = 18/96 (18%)
Query 8 TSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGAL 67
S L+LA+G D ++++WE T L L H D VRD+SF P G+ L
Sbjct 121 VSCLILATGLNDGQIKIWEVQ-----TGLLLLNLSGHQDVVRDLSFTPSGS--------L 167
Query 68 LLASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQ 103
+L S S DKT+++W +H +Q Q L Q
Sbjct 168 ILVSASRDKTLRIW-----DLNKHGKQIQVLSGHLQ 198
> hsa:55884 WSB2, MGC10210, SBA2; WD repeat and SOCS box containing
2; K10342 WD repeat and SOCS box-containing protein 2
Length=404
Score = 45.4 bits (106), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 47/96 (48%), Gaps = 18/96 (18%)
Query 8 TSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGAL 67
S L+LA+G D ++++WE T L L H D VRD+SF P G+ L
Sbjct 121 VSCLVLATGLNDGQIKIWEVQ-----TGLLLLNLSGHQDVVRDLSFTPSGS--------L 167
Query 68 LLASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQ 103
+L S S DKT+++W +H +Q Q L Q
Sbjct 168 ILVSASRDKTLRIW-----DLNKHGKQIQVLSGHLQ 198
> dre:323226 wsb1, wu:fb92g11, zgc:63905; WD repeat and SOCS box-containing
1; K10341 WD repeat and SOCS box-containing protein
1
Length=423
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 39/71 (54%), Gaps = 13/71 (18%)
Query 11 LLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLA 70
LLLA+G + R+++W+ K L HTD VRD++F P G+ L+L
Sbjct 142 LLLATGLNNGRIKIWDVYTGKLLLNL-----MDHTDIVRDLTFAPDGS--------LVLV 188
Query 71 SCSEDKTVKLW 81
S S DKT+++W
Sbjct 189 SASRDKTLRVW 199
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 35/80 (43%), Gaps = 17/80 (21%)
Query 10 GLLLASGGADNRVRLWECDASKQWTEL--------PALAGEHHTDWVRDVSFRPQGASSF 61
G LLA+ D RV +W+ + EL P AG + WVR V+F G
Sbjct 269 GALLATASYDTRVIVWDPHTATVLLELGHLFPPPSPIFAGGANDRWVRSVAFCHDGRH-- 326
Query 62 VVPGALLLASCSEDKTVKLW 81
+AS ++D+ V+ W
Sbjct 327 -------IASVTDDRLVRFW 339
> xla:379573 poc1a, MGC69111, pix2, wdr51a; POC1 centriolar protein
homolog A (Chlamydomonas)
Length=399
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 37/79 (46%), Gaps = 14/79 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
SVD SG L+AS D VRLW + T A HT VR VSF G S
Sbjct 23 SVDFSPSGHLIASASRDKTVRLWVPSVKGESTAFKA-----HTGTVRSVSFSGDGQS--- 74
Query 63 VPGALLLASCSEDKTVKLW 81
L + S+DKT+K+W
Sbjct 75 ------LVTASDDKTIKVW 87
Score = 32.7 bits (73), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 39/160 (24%), Positives = 60/160 (37%), Gaps = 25/160 (15%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
G L+ S D ++LW+ S++ + + H +V V F P G +
Sbjct 114 GRLIVSASDDKTIKLWD-KTSRECIQ----SFCEHGGFVNFVDFHPSGT---------CI 159
Query 70 ASCSEDKTVKLWIGEPPQQQQHQQQQ----QQLQQQQQQDILMNNPSPSQYK------WR 119
A+ + D TVK+W + QH Q L + L+ + S K R
Sbjct 160 AAAATDNTVKVWDIRMNKLIQHYQVHSGVVNSLSFHPSGNYLITASNDSTLKVLDLLEGR 219
Query 120 LLQTLV-LDAPAWRVTWSSSGALLAVACGDSAVLLLRENM 158
LL TL P V +S G A D V++ + N
Sbjct 220 LLYTLHGHQGPVTCVKFSREGDFFASGGSDEQVMVWKTNF 259
Score = 32.0 bits (71), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 9/49 (18%)
Query 34 TELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASCSEDKTVKLWI 82
T++ A H D + V F P G L+AS S DKTV+LW+
Sbjct 7 TQMRAYRFVGHKDAILSVDFSPSGH---------LIASASRDKTVRLWV 46
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 32/77 (41%), Gaps = 14/77 (18%)
Query 4 VDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVV 63
VD SG +A+ DN V++W+ +K H+ V +SF P G
Sbjct 150 VDFHPSGTCIAAAATDNTVKVWDIRMNKLIQHYQV-----HSGVVNSLSFHPSGN----- 199
Query 64 PGALLLASCSEDKTVKL 80
L + S D T+K+
Sbjct 200 ----YLITASNDSTLKV 212
> xla:380049 wsb1, MGC53424; WD repeat and SOCS box containing
1; K10341 WD repeat and SOCS box-containing protein 1
Length=244
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 13/71 (18%)
Query 11 LLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLA 70
LLLA+G ++ R+++W+ K L HT+ VRD++F P G+ L+L
Sbjct 141 LLLATGLSNGRIKIWDVYTGKLLLNL-----MDHTEVVRDLTFAPDGS--------LILV 187
Query 71 SCSEDKTVKLW 81
S S DKT+++W
Sbjct 188 SASRDKTLRVW 198
> hsa:26118 WSB1, SWIP1, WSB-1; WD repeat and SOCS box containing
1; K10341 WD repeat and SOCS box-containing protein 1
Length=421
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 13/71 (18%)
Query 11 LLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLA 70
LLLA+G + R+++W+ K L HT+ VRD++F P G+ L+L
Sbjct 141 LLLATGLNNGRIKIWDVYTGKLLLNL-----VDHTEVVRDLTFAPDGS--------LILV 187
Query 71 SCSEDKTVKLW 81
S S DKT+++W
Sbjct 188 SASRDKTLRVW 198
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 36/87 (41%), Gaps = 17/87 (19%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTEL--------PALAGEHHTDWVRDVSFR 54
+ D G LLA+ D RV +W+ E P AG + WVR VSF
Sbjct 261 ACDFSPDGALLATASYDTRVYIWDPHNGDILMEFGHLFPPPTPIFAGGANDRWVRSVSFS 320
Query 55 PQGASSFVVPGALLLASCSEDKTVKLW 81
G L +AS ++DK V+ W
Sbjct 321 HDG---------LHVASLADDKMVRFW 338
> ath:AT4G32551 LUG; LUG (LEUNIG); protein binding / protein heterodimerization/
transcription repressor
Length=931
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/74 (36%), Positives = 35/74 (47%), Gaps = 14/74 (18%)
Query 8 TSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGAL 67
+ G +LAS G D + LW D K T L E HT + D+ F P L
Sbjct 661 SDGKMLASAGHDKKAVLWYTDTMKPKTTL-----EEHTAMITDIRFSPS---------QL 706
Query 68 LLASCSEDKTVKLW 81
LA+ S DKTV++W
Sbjct 707 RLATSSFDKTVRVW 720
> mmu:67561 Wdr48, 8430408H12Rik, mKIAA1449; WD repeat domain
48
Length=676
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 35/69 (50%), Gaps = 11/69 (15%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
L + G D+ +R+W + KQ + ++ EHHTDWV DV G + L S
Sbjct 45 LFTAGRDSIIRIWSVNQHKQDPYIASM--EHHTDWVNDVVLCCNGKT---------LISA 93
Query 73 SEDKTVKLW 81
S D TVK+W
Sbjct 94 SSDTTVKVW 102
> ath:AT2G05720 transducin family protein / WD-40 repeat family
protein
Length=276
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 37/81 (45%), Gaps = 13/81 (16%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
SVD +G LASGG DN+ R+W+ K +PA H + V V + PQ
Sbjct 177 SVDFSPNGYHLASGGEDNQCRIWDLRMRKLLYIIPA-----HVNLVSQVKYEPQ------ 225
Query 63 VPGALLLASCSEDKTVKLWIG 83
LA+ S D V +W G
Sbjct 226 --ERYFLATASHDMNVNIWSG 244
> xla:446524 taf5l, MGC80243; TAF5-like RNA polymerase II, p300/CBP-associated
factor (PCAF)-associated factor, 65kDa; K03130
transcription initiation factor TFIID subunit 5
Length=587
Score = 42.7 bits (99), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 38/73 (52%), Gaps = 14/73 (19%)
Query 9 SGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALL 68
+G LAS G D R++LW+ + Q+ EL HTD + ++F P + L
Sbjct 477 NGKYLASAGEDQRLKLWDLASGTQYKELRG-----HTDNISSLTFSPDSS---------L 522
Query 69 LASCSEDKTVKLW 81
+AS S D +V++W
Sbjct 523 IASASMDNSVRVW 535
Score = 32.3 bits (72), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 33/69 (47%), Gaps = 14/69 (20%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
LA+G +D VRLW +++Q + G H V ++F P G LAS
Sbjct 439 LATGSSDKTVRLW---STQQGNSVRLFTG--HRGPVLTLAFSPNGK---------YLASA 484
Query 73 SEDKTVKLW 81
ED+ +KLW
Sbjct 485 GEDQRLKLW 493
Score = 30.0 bits (66), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 32/74 (43%), Gaps = 16/74 (21%)
Query 8 TSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGAL 67
+SGLL S D +R W ++ +T G + W DVS P +L
Sbjct 352 SSGLL--SCSEDTSIRYWNLES---YTNTVLYQGHTYPVWDLDVS-----------PCSL 395
Query 68 LLASCSEDKTVKLW 81
AS S D+T +LW
Sbjct 396 FFASASHDRTGRLW 409
> dre:386932 poc1b, TUWD12, fl36w17, wdr51b, wu:fl36w17, zgc:63538;
POC1 centriolar protein homolog B (Chlamydomonas)
Length=490
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 36/79 (45%), Gaps = 14/79 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
SV+ G L + D V++W + K L HT+WVR F P G
Sbjct 107 SVNFSRDGQRLVTASDDKSVKVWGVERKKFLYSL-----NRHTNWVRCARFSPDGR---- 157
Query 63 VPGALLLASCSEDKTVKLW 81
L+ASC +D+TV+LW
Sbjct 158 -----LIASCGDDRTVRLW 171
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 42/156 (26%), Positives = 57/156 (36%), Gaps = 47/156 (30%)
Query 4 VDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVV 63
VD +SG +AS GADN +++W+ +K H V SF P G
Sbjct 192 VDFNSSGTCIASSGADNTIKIWDIRTNKLIQHYKV-----HNAGVNCFSFHPSGN----- 241
Query 64 PGALLLASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQT 123
L S S D T+K+ D+L + RL+ T
Sbjct 242 ----YLISGSSDSTIKIL-----------------------DLL---------EGRLIYT 265
Query 124 LV-LDAPAWRVTWSSSGALLAVACGDSAVLLLRENM 158
L P VT+S G L A DS VL+ + N
Sbjct 266 LHGHKGPVLTVTFSRDGDLFASGGADSQVLMWKTNF 301
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 41/171 (23%), Positives = 64/171 (37%), Gaps = 25/171 (14%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
LA+G D + +W + A HTD + V+F P G+ L+AS
Sbjct 33 LATGSCDKSLMIWNLAPKAR-----AFRFVGHTDVITGVNFAPSGS---------LVASS 78
Query 73 SEDKTVKLWI----GEPPQQQQHQQQQQQLQ-QQQQQDILMNNPSPSQYKWRLLQTLVLD 127
S D+TV+LW GE + H + + + Q ++ + S W + + L
Sbjct 79 SRDQTVRLWTPSIKGESTVFKAHTASVRSVNFSRDGQRLVTASDDKSVKVWGVERKKFLY 138
Query 128 APAWRVTW------SSSGALLAVACGDSAVLLLRENMHGTWELVTDLAEKA 172
+ W S G L+A D V L + H + TD A
Sbjct 139 SLNRHTNWVRCARFSPDGRLIASCGDDRTVRLWDTSSHQCTNIFTDYGGSA 189
> hsa:57599 WDR48, DKFZp686G1794, KIAA1449, P80, UAF1; WD repeat
domain 48
Length=677
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 35/69 (50%), Gaps = 11/69 (15%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
L + G D+ +R+W + KQ + ++ EHHTDWV D+ G + L S
Sbjct 45 LFTAGRDSIIRIWSVNQHKQDPYIASM--EHHTDWVNDIVLCCNGKT---------LISA 93
Query 73 SEDKTVKLW 81
S D TVK+W
Sbjct 94 SSDTTVKVW 102
> dre:334682 wdr48, fk50c12, wd48, wu:fd19a04, wu:fk50c12, zgc:66229;
WD repeat domain 48
Length=677
Score = 42.4 bits (98), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 35/69 (50%), Gaps = 11/69 (15%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
L + G D+ +R+W + KQ + ++ EHHTDWV D+ G + L S
Sbjct 45 LFTAGRDSIIRIWSVNQHKQDPYIASM--EHHTDWVNDIILCCNGKT---------LISA 93
Query 73 SEDKTVKLW 81
S D TVK+W
Sbjct 94 SSDTTVKVW 102
> dre:100333793 WD repeat-containing protein 48-like
Length=677
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 35/69 (50%), Gaps = 11/69 (15%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
L + G D+ +R+W + KQ + ++ EHHTDWV D+ G + L S
Sbjct 45 LFTAGRDSIIRIWSVNQHKQDPYIASM--EHHTDWVNDIILCCNGKT---------LISA 93
Query 73 SEDKTVKLW 81
S D TVK+W
Sbjct 94 SSDTTVKVW 102
> tgo:TGME49_119570 WD-40 repeat protein, putative (EC:2.7.11.7)
Length=552
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 38/75 (50%), Gaps = 14/75 (18%)
Query 7 RTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGA 66
R +GL+L S D +R+W+ + Q T + G H +WVR V P G+
Sbjct 427 RVAGLVLFSASRDRTLRMWD---AMQGTTMHVFVG--HDNWVRRVVLHPGGSH------- 474
Query 67 LLLASCSEDKTVKLW 81
+ SCS+D++++ W
Sbjct 475 --IISCSDDRSIRCW 487
> ath:AT2G41500 EMB2776; nucleotide binding; K12662 U4/U6 small
nuclear ribonucleoprotein PRP4
Length=554
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 37/81 (45%), Gaps = 13/81 (16%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
SV+ +G LASGG DN+ R+W+ K +PA H + V V + PQ
Sbjct 428 SVNFSPNGYHLASGGEDNQCRIWDLRMRKSLYIIPA-----HANLVSQVKYEPQ------ 476
Query 63 VPGALLLASCSEDKTVKLWIG 83
LA+ S D V +W G
Sbjct 477 --EGYFLATASYDMKVNIWSG 495
> mmu:70235 Poc1a, 2510040D07Rik, Wdr51a; POC1 centriolar protein
homolog A (Chlamydomonas)
Length=405
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 38/80 (47%), Gaps = 14/80 (17%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
+ V+ SG LLASG D VR+W + + T A HT VR V F G S
Sbjct 65 TCVNFSPSGHLLASGSRDKTVRIWVPNVKGESTVFRA-----HTATVRSVHFCSDGQS-- 117
Query 62 VVPGALLLASCSEDKTVKLW 81
L + S+DKTVK+W
Sbjct 118 -------LVTASDDKTVKVW 130
Score = 37.0 bits (84), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 44/160 (27%), Positives = 59/160 (36%), Gaps = 25/160 (15%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
G L+ S D V+LW+ K E EH +V V F P G +
Sbjct 157 GRLIVSASDDKTVKLWD----KTSRECIHSYCEH-GGFVTYVDFHPSGT---------CI 202
Query 70 ASCSEDKTVKLWIGEPPQQQQHQQQQ----QQLQQQQQQDILMNNPSPSQYKW------R 119
A+ D TVK+W + QH Q L + L+ S S K R
Sbjct 203 AAAGMDNTVKVWDARTHRLLQHYQLHSAAVNALSFHPSGNYLITASSDSTLKILDLMEGR 262
Query 120 LLQTLV-LDAPAWRVTWSSSGALLAVACGDSAVLLLRENM 158
LL TL PA V +S +G A D V++ + N
Sbjct 263 LLYTLHGHQGPATTVAFSRTGEYFASGGSDEQVMVWKSNF 302
Score = 36.2 bits (82), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 33/79 (41%), Gaps = 14/79 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
SV + G L + D V++W + L H +WVR F P G
Sbjct 108 SVHFCSDGQSLVTASDDKTVKVWSTHRQRFLFSL-----TQHINWVRCAKFSPDGR---- 158
Query 63 VPGALLLASCSEDKTVKLW 81
L+ S S+DKTVKLW
Sbjct 159 -----LIVSASDDKTVKLW 172
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 31/70 (44%), Gaps = 14/70 (20%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
LASG D+ + +W + H D V V+F P G LLAS
Sbjct 34 LASGSMDSTLMIWHMKPQSRAYRFTG-----HKDAVTCVNFSPSGH---------LLASG 79
Query 73 SEDKTVKLWI 82
S DKTV++W+
Sbjct 80 SRDKTVRIWV 89
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 18/79 (22%)
Query 4 VDGRTSGLLLASGGADNRVRLWECDASK--QWTELPALAGEHHTDWVRDVSFRPQGASSF 61
VD SG +A+ G DN V++W+ + Q +L H+ V +SF P G
Sbjct 193 VDFHPSGTCIAAAGMDNTVKVWDARTHRLLQHYQL-------HSAAVNALSFHPSGN--- 242
Query 62 VVPGALLLASCSEDKTVKL 80
L + S D T+K+
Sbjct 243 ------YLITASSDSTLKI 255
> xla:399199 pafah1b1, lis2, mdcr, mds, pafah, pafah1b1-b; platelet-activating
factor acetylhydrolase 1b, regulatory subunit
1 (45kDa) (EC:3.1.1.47); K01062 1-alkyl-2-acetylglycerophosphocholine
esterase [EC:3.1.1.47]
Length=410
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 34/72 (47%), Gaps = 14/72 (19%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
G L SG D +++W+ L L G H +WVR V F P G +
Sbjct 308 GPFLLSGSRDKTIKMWDISIGMC---LMTLVG--HDNWVRGVQFHP---------GGKFI 353
Query 70 ASCSEDKTVKLW 81
SC++DKT+++W
Sbjct 354 LSCADDKTIRIW 365
Score = 36.6 bits (83), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 33/70 (47%), Gaps = 14/70 (20%)
Query 12 LLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
++ + D +++W+ + L HTD V+D+SF G LLAS
Sbjct 122 VMVTASEDATIKVWDYETGDFERTLKG-----HTDSVQDISFDHSGK---------LLAS 167
Query 72 CSEDKTVKLW 81
CS D T+KLW
Sbjct 168 CSADMTIKLW 177
Score = 33.9 bits (76), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 38/81 (46%), Gaps = 14/81 (17%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
SSV +G + S D +++WE + G H +WVR V RP +
Sbjct 196 SSVAIMPNGDHIVSASRDKTIKMWEVQTGYC---VKTFTG--HREWVRMV--RPNQDGT- 247
Query 62 VVPGALLLASCSEDKTVKLWI 82
L+ASCS D+TV++W+
Sbjct 248 ------LIASCSNDQTVRVWV 262
Score = 33.1 bits (74), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 33/73 (45%), Gaps = 14/73 (19%)
Query 9 SGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALL 68
SG LLAS AD ++LW+ + L + G H V V+ P G
Sbjct 161 SGKLLASCSADMTIKLWDFQG---FECLRTMHGHDHN--VSSVAIMPNGDH--------- 206
Query 69 LASCSEDKTVKLW 81
+ S S DKT+K+W
Sbjct 207 IVSASRDKTIKMW 219
> dre:406285 chaf1b, wu:fd07c09, wu:fe36g06, wu:fv38g09, zgc:56096;
chromatin assembly factor 1, subunit B; K10751 chromatin
assembly factor 1 subunit B
Length=236
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 39/156 (25%), Positives = 67/156 (42%), Gaps = 28/156 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGE--HHTDWVRDVSFRPQGASS 60
S DG+T LA+ G D VR+W D + HT V V F P
Sbjct 24 SGDGKTQ--RLATAGVDTTVRMWRVDKGPDGKAVVEFLSNLARHTKAVNVVRFSPT---- 77
Query 61 FVVPGALLLASCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRL 120
A +LAS +D + LW + +++ +Q Q+++D +N S W +
Sbjct 78 -----AEVLASGGDDAAILLW-----KLNDNKEPEQTPTFQEEEDAQLNKES-----WSV 122
Query 121 LQTL---VLDAPAWRVTWSSSGALLAVACGDSAVLL 153
++TL + D + ++W+S G +A D+ ++
Sbjct 123 VKTLRGHIED--VYDISWTSDGNFMASGSVDNTAIM 156
> hsa:8208 CHAF1B, CAF-1, CAF-IP60, CAF1, CAF1A, CAF1P60, MPHOSPH7,
MPP7; chromatin assembly factor 1, subunit B (p60); K10751
chromatin assembly factor 1 subunit B
Length=559
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 34/146 (23%), Positives = 62/146 (42%), Gaps = 27/146 (18%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGE--HHTDWVRDVSFRPQGASSFVVPGALLLA 70
LAS G D VR+W+ + + HT V V F P G +LA
Sbjct 32 LASAGVDTNVRIWKVEKGPDGKAIVEFLSNLARHTKAVNVVRFSPTGE---------ILA 82
Query 71 SCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTL---VLD 127
S +D + LW + ++ +Q+ Q + + +N + W +++TL + D
Sbjct 83 SGGDDAVILLW------KVNDNKEPEQIAFQDEDEAQLN-----KENWTVVKTLRGHLED 131
Query 128 APAWRVTWSSSGALLAVACGDSAVLL 153
+ + W++ G L+A A D+ ++
Sbjct 132 --VYDICWATDGNLMASASVDNTAII 155
> dre:795104 ciao1, MGC55911, MGC77394, wdr39, zgc:55911, zgc:77394;
cytosolic iron-sulfur protein assembly 1 homolog (S.
cerevisiae)
Length=330
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/148 (25%), Positives = 58/148 (39%), Gaps = 37/148 (25%)
Query 12 LLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
LLAS DN++ +++ + W L G T W ++F P+G LAS
Sbjct 164 LLASASYDNKICIYK-EEDDDWECRATLEGHESTVW--SLTFDPEGRR---------LAS 211
Query 72 CSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTL--VLDAP 129
CS+D+TVK+W ++ D S W+ + TL
Sbjct 212 CSDDRTVKIW-----------------KESTTGD------GSSDESWKCICTLSGFHGRT 248
Query 130 AWRVTWSSSGALLAVACGDSAVLLLREN 157
+ + W LA ACGD V + E+
Sbjct 249 IYDIAWCRLTGALATACGDDGVRVFSED 276
Score = 35.0 bits (79), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 44/182 (24%), Positives = 77/182 (42%), Gaps = 34/182 (18%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
G LAS D +W+ + + L L G H + V+ V++ P G+ LL
Sbjct 73 GKYLASASFDATTCIWK-KTDEDFECLTVLEG--HENEVKCVAWAPSGS---------LL 120
Query 70 ASCSEDKTVKLW-IGEPPQQQ------QHQQQQQQLQQQQQQDILMNNPSPSQ---YK-- 117
A+CS DK+V +W + E + + H Q + + Q++L + ++ YK
Sbjct 121 ATCSRDKSVWIWEVDEEDEYECLSVVNSHTQDVKHVVWHPTQELLASASYDNKICIYKEE 180
Query 118 ---WRLLQTLV-LDAPAWRVTWSSSGALLAVACGDSAVLLLRENMHG------TWELVTD 167
W TL ++ W +T+ G LA D V + +E+ G +W+ +
Sbjct 181 DDDWECRATLEGHESTVWSLTFDPEGRRLASCSDDRTVKIWKESTTGDGSSDESWKCICT 240
Query 168 LA 169
L+
Sbjct 241 LS 242
> sce:YCR072C RSA4; Rsa4p; K14855 ribosome assembly protein 4
Length=515
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 35/69 (50%), Gaps = 14/69 (20%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
+ +G DN R+W+CD T+ P + H +WV VS+ P G ++A+
Sbjct 159 MVTGAGDNTARIWDCD-----TQTPMHTLKGHYNWVLCVSWSPDGE---------VIATG 204
Query 73 SEDKTVKLW 81
S D T++LW
Sbjct 205 SMDNTIRLW 213
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 26/73 (35%), Positives = 42/73 (57%), Gaps = 8/73 (10%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGAL-L 68
G ++A+G DN +RLW+ + + + AL G H+ W+ +S+ P V PG+
Sbjct 198 GEVIATGSMDNTIRLWDPKSGQCLGD--ALRG--HSKWITSLSWEPI---HLVKPGSKPR 250
Query 69 LASCSEDKTVKLW 81
LAS S+D T+K+W
Sbjct 251 LASSSKDGTIKIW 263
> mmu:110749 Chaf1b, 2600017H24Rik, C76145, CAF-I_p60, CAF-Ip60,
CAF1, CAF1A, CAF1P60, MPHOSPH7; chromatin assembly factor
1, subunit B (p60); K10751 chromatin assembly factor 1 subunit
B
Length=572
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/146 (23%), Positives = 63/146 (43%), Gaps = 27/146 (18%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGE--HHTDWVRDVSFRPQGASSFVVPGALLLA 70
LAS G D VR+W+ + + HT V V F P G +LA
Sbjct 32 LASAGVDTAVRIWKLERGPDGKAIVEFLSNLARHTKAVNVVRFSPTGE---------ILA 82
Query 71 SCSEDKTVKLWIGEPPQQQQHQQQQQQLQQQQQQDILMNNPSPSQYKWRLLQTL---VLD 127
S +D + LW + ++ +Q+ Q +++ +N + W +++TL + D
Sbjct 83 SGGDDAVILLW------KMNDSKEPEQIAFQDEEEAQLN-----KENWTVVKTLRGHLED 131
Query 128 APAWRVTWSSSGALLAVACGDSAVLL 153
+ + W++ G L+ A D+ V++
Sbjct 132 --VYDICWATDGNLMTSASVDNTVII 155
Score = 28.9 bits (63), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 38/91 (41%), Gaps = 29/91 (31%)
Query 9 SGLLLASGGADNRVRLWECDASKQ------------------WTELPALAGEHHTDWVRD 50
+G +LASGG D + LW+ + SK+ WT + L G H + V D
Sbjct 77 TGEILASGGDDAVILLWKMNDSKEPEQIAFQDEEEAQLNKENWTVVKTLRG--HLEDVYD 134
Query 51 VSFRPQGASSFVVPGALLLASCSEDKTVKLW 81
+ + G L+ S S D TV +W
Sbjct 135 ICWATDGN---------LMTSASVDNTVIIW 156
> sce:YIL046W MET30, ZRG11; F-box protein containing five copies
of the WD40 motif, controls cell cycle function, sulfur metabolism,
and methionine biosynthesis as part of the ubiquitin
ligase complex; interacts with and regulates Met4p, localizes
within the nucleus; K10259 F-box and WD-40 domain protein
MET30
Length=640
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 23/70 (32%), Positives = 36/70 (51%), Gaps = 15/70 (21%)
Query 12 LLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
++ SG AD V++W ++ +T L G HT+WV V P+ S F S
Sbjct 394 VIVSGSADKTVKVWHVESRTCYT----LRG--HTEWVNCVKLHPKSFSCF---------S 438
Query 72 CSEDKTVKLW 81
CS+D T+++W
Sbjct 439 CSDDTTIRMW 448
> ath:AT2G32700 LUH; WD-40 repeat family protein
Length=785
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/77 (35%), Positives = 36/77 (46%), Gaps = 14/77 (18%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
G LLAS G D +V +W + + Q P E H + DV FRP + L
Sbjct 520 GKLLASAGHDKKVFIWNME-TLQVESTP----EEHAHIITDVRFRPN---------STQL 565
Query 70 ASCSEDKTVKLWIGEPP 86
A+ S DKT+K+W P
Sbjct 566 ATSSFDKTIKIWDASDP 582
> ath:AT4G29830 VIP3; VIP3 (vernalization independence 3); nucleotide
binding / protein binding; K12602 WD repeat-containing
protein 61
Length=321
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 39/79 (49%), Gaps = 8/79 (10%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
SVD G +A+G +D VRLW+ + +H D V V+FRP G +
Sbjct 248 SVDASPDGGAIATGSSDRTVRLWDLKMRAAIQTM-----SNHNDQVWSVAFRPPGGTGV- 301
Query 63 VPGALLLASCSEDKTVKLW 81
A LAS S+DK+V L+
Sbjct 302 --RAGRLASVSDDKSVSLY 318
Score = 33.1 bits (74), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 39/81 (48%), Gaps = 18/81 (22%)
Query 1 YSSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASS 60
+S VD R +L SG D V + + + T L +++G HT WV V P G +
Sbjct 208 FSPVDPR----VLFSGSDDGHVNMHDAEGK---TLLGSMSG--HTSWVLSVDASPDGGA- 257
Query 61 FVVPGALLLASCSEDKTVKLW 81
+A+ S D+TV+LW
Sbjct 258 --------IATGSSDRTVRLW 270
> dre:394247 pafah1b1b, Lis1a; platelet-activating factor acetylhydrolase,
isoform Ib, alpha subunit b; K01062 1-alkyl-2-acetylglycerophosphocholine
esterase [EC:3.1.1.47]
Length=410
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 36/72 (50%), Gaps = 14/72 (19%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
G L SG D +++W+ L L G H +WVR V F P G FVV
Sbjct 308 GPFLLSGSRDKTIKMWDISTGMC---LMTLVG--HDNWVRGVLFHPGG--RFVV------ 354
Query 70 ASCSEDKTVKLW 81
SC++DKT+++W
Sbjct 355 -SCADDKTLRIW 365
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 25/70 (35%), Positives = 34/70 (48%), Gaps = 14/70 (20%)
Query 12 LLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
L+ S D +++W+ +A L HTD V+D+SF G LLAS
Sbjct 122 LMVSASEDATIKVWDYEAGDFERTLKG-----HTDSVQDISFDQTGK---------LLAS 167
Query 72 CSEDKTVKLW 81
CS D T+KLW
Sbjct 168 CSADMTIKLW 177
Score = 33.1 bits (74), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 14/81 (17%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
SSV +G + S D +++WE + G H +WVR V RP +
Sbjct 196 SSVAIMPNGDHIVSASRDKTMKMWEVATGYC---VKTFTG--HREWVRMV--RPNQDGT- 247
Query 62 VVPGALLLASCSEDKTVKLWI 82
LLASCS D+TV++W+
Sbjct 248 ------LLASCSNDQTVRVWV 262
Score = 30.4 bits (67), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 14/73 (19%)
Query 9 SGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALL 68
+G LLAS AD ++LW+ + + + G H V V+ P G
Sbjct 161 TGKLLASCSADMTIKLWDFQG---FECIRTMHGHDHN--VSSVAIMPNGDH--------- 206
Query 69 LASCSEDKTVKLW 81
+ S S DKT+K+W
Sbjct 207 IVSASRDKTMKMW 219
> dre:406291 katnb1, wu:fj32f02, wu:fj65h01, zgc:56071; katanin
p80 (WD repeat containing) subunit B 1
Length=694
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 28/69 (40%), Positives = 33/69 (47%), Gaps = 14/69 (20%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
LAS D+ V+LW+ A K TE + HT V V F P LLAS
Sbjct 162 LASASDDSTVKLWDLIAGKMITEFTS-----HTSAVNVVQFHPN---------EYLLASG 207
Query 73 SEDKTVKLW 81
S D+TVKLW
Sbjct 208 SADRTVKLW 216
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 39/80 (48%), Gaps = 14/80 (17%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
SS+D G LASG D+ ++LW D ++ + HT VR ++F P G
Sbjct 109 SSLDFHPMGEYLASGSVDSNIKLW--DVRRKGC---VFRYKGHTQAVRCLAFSPDGK--- 160
Query 62 VVPGALLLASCSEDKTVKLW 81
LAS S+D TVKLW
Sbjct 161 ------WLASASDDSTVKLW 174
Score = 31.6 bits (70), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 5/47 (10%)
Query 12 LLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGA 58
LLASG AD V+LW+ + ++ + + GE T VR V F P G+
Sbjct 203 LLASGSADRTVKLWDLE---KFNMIGSSEGE--TGVVRSVLFNPDGS 244
Score = 31.2 bits (69), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 35/66 (53%), Gaps = 7/66 (10%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
S V G++SG LLA+GG D RV +W A + + +L G HT V + F +
Sbjct 25 SLVLGKSSGRLLATGGEDCRVNIW---AVSKPNCIMSLTG--HTSAVGCIQF--NSSEER 77
Query 62 VVPGAL 67
VV G+L
Sbjct 78 VVAGSL 83
> hsa:25886 POC1A, DKFZp434C245, MGC131902, PIX2, WDR51A; POC1
centriolar protein homolog A (Chlamydomonas)
Length=359
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 37/80 (46%), Gaps = 14/80 (17%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
+ V+ SG LLASG D VR+W + + T A HT VR V F G S
Sbjct 65 TCVNFSPSGHLLASGSRDKTVRIWVPNVKGESTVFRA-----HTATVRSVHFCSDGQS-- 117
Query 62 VVPGALLLASCSEDKTVKLW 81
+ S+DKTVK+W
Sbjct 118 -------FVTASDDKTVKVW 130
Score = 37.0 bits (84), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 44/160 (27%), Positives = 59/160 (36%), Gaps = 25/160 (15%)
Query 10 GLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLL 69
G L+ S D V+LW+ K E EH +V V F P G +
Sbjct 157 GRLIVSASDDKTVKLWD----KSSRECVHSYCEH-GGFVTYVDFHPSGT---------CI 202
Query 70 ASCSEDKTVKLWIGEPPQQQQHQQQQQQ----LQQQQQQDILMNNPSPSQYKW------R 119
A+ D TVK+W + QH Q L + L+ S S K R
Sbjct 203 AAAGMDNTVKVWDVRTHRLLQHYQLHSAAVNGLSFHPSGNYLITASSDSTLKILDLMEGR 262
Query 120 LLQTLV-LDAPAWRVTWSSSGALLAVACGDSAVLLLRENM 158
LL TL PA V +S +G A D V++ + N
Sbjct 263 LLYTLHGHQGPATTVAFSRTGEYFASGGSDEQVMVWKSNF 302
Score = 33.9 bits (76), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 35/81 (43%), Gaps = 14/81 (17%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSF 61
+ VD + LASG D+ + +W + H D V V+F P G
Sbjct 23 TCVDFSINTKQLASGSMDSCLMVWHMKPQSRAYRFTG-----HKDAVTCVNFSPSGH--- 74
Query 62 VVPGALLLASCSEDKTVKLWI 82
LLAS S DKTV++W+
Sbjct 75 ------LLASGSRDKTVRIWV 89
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 18/79 (22%)
Query 4 VDGRTSGLLLASGGADNRVRLWECDASK--QWTELPALAGEHHTDWVRDVSFRPQGASSF 61
VD SG +A+ G DN V++W+ + Q +L H+ V +SF P G
Sbjct 193 VDFHPSGTCIAAAGMDNTVKVWDVRTHRLLQHYQL-------HSAAVNGLSFHPSGN--- 242
Query 62 VVPGALLLASCSEDKTVKL 80
L + S D T+K+
Sbjct 243 ------YLITASSDSTLKI 255
> mmu:74187 Katnb1, 2410003J24Rik, KAT; katanin p80 (WD40-containing)
subunit B 1
Length=658
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 32/69 (46%), Gaps = 14/69 (20%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
LAS D+ V+LW+ A K +E P HT V V F P LLAS
Sbjct 162 LASAADDHTVKLWDLTAGKMMSEFPG-----HTGPVNVVEFHPN---------EYLLASG 207
Query 73 SEDKTVKLW 81
S D+T++ W
Sbjct 208 SSDRTIRFW 216
Score = 33.1 bits (74), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 35/79 (44%), Gaps = 14/79 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
S+D G +ASG D ++LW D ++ H+ VR + F P G
Sbjct 110 SLDFHPYGEFVASGSQDTNIKLW--DIRRKGC---VFRYRGHSQAVRCLRFSPDGK---- 160
Query 63 VPGALLLASCSEDKTVKLW 81
LAS ++D TVKLW
Sbjct 161 -----WLASAADDHTVKLW 174
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 15/27 (55%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECD 28
S V G+ SG LLA+GG D RV LW +
Sbjct 25 SLVLGKASGRLLATGGDDCRVNLWSIN 51
> hsa:10300 KATNB1, KAT; katanin p80 (WD repeat containing) subunit
B 1
Length=655
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 32/69 (46%), Gaps = 14/69 (20%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFVVPGALLLASC 72
LAS D+ V+LW+ A K +E P HT V V F P LLAS
Sbjct 162 LASAADDHTVKLWDLTAGKMMSEFPG-----HTGPVNVVEFHPN---------EYLLASG 207
Query 73 SEDKTVKLW 81
S D+T++ W
Sbjct 208 SSDRTIRFW 216
Score = 33.1 bits (74), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 25/79 (31%), Positives = 35/79 (44%), Gaps = 14/79 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
S+D G +ASG D ++LW D ++ H+ VR + F P G
Sbjct 110 SLDFHPYGEFVASGSQDTNIKLW--DIRRKGC---VFRYRGHSQAVRCLRFSPDGK---- 160
Query 63 VPGALLLASCSEDKTVKLW 81
LAS ++D TVKLW
Sbjct 161 -----WLASAADDHTVKLW 174
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 15/27 (55%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 2 SSVDGRTSGLLLASGGADNRVRLWECD 28
S V G+ SG LLA+GG D RV LW +
Sbjct 25 SLVLGKASGRLLATGGDDCRVNLWSIN 51
> cel:Y53C12B.1 hypothetical protein; K14555 U3 small nucleolar
RNA-associated protein 13
Length=793
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 30/79 (37%), Positives = 37/79 (46%), Gaps = 12/79 (15%)
Query 4 VDGRTSGLLLASGGADNRVRLWECDASK-QWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
VD S L+A+GG D V+LW+ D K Q L+G H V DV F
Sbjct 483 VDISESDALIATGGMDKLVKLWQVDTHKMQLGIAGTLSG--HRRGVGDVKFAKNSHK--- 537
Query 63 VPGALLLASCSEDKTVKLW 81
LASCS D T+K+W
Sbjct 538 ------LASCSGDMTIKIW 550
Score = 32.0 bits (71), Expect = 1.1, Method: Composition-based stats.
Identities = 24/74 (32%), Positives = 32/74 (43%), Gaps = 11/74 (14%)
Query 12 LLASGGADNRVRLWECDASKQ----WTELPALAGEHHTDWVRDVSFRPQGASSFVVPGAL 67
LLAS DN + W S + T +P A H + V ++ G + F
Sbjct 375 LLASCSKDNSIIFWRLVTSPENDSCSTLVPVAAATGHANTVTALAISNTGRAPF------ 428
Query 68 LLASCSEDKTVKLW 81
LAS S D T+KLW
Sbjct 429 -LASVSTDCTIKLW 441
> cel:T10F2.4 hypothetical protein; K10599 pre-mRNA-processing
factor 19 [EC:6.3.2.19]
Length=492
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 14/79 (17%)
Query 3 SVDGRTSGLLLASGGADNRVRLWECDASKQWTELPALAGEHHTDWVRDVSFRPQGASSFV 62
S++ GL+ +G AD V++W+ K T A G HT VR ++F G
Sbjct 343 SIEFHPDGLIFGTGAADAVVKIWDL---KNQTVAAAFPG--HTAAVRSIAFSENG----- 392
Query 63 VPGALLLASCSEDKTVKLW 81
LA+ SED VKLW
Sbjct 393 ----YYLATGSEDGEVKLW 407
> dre:777754 zgc:153492
Length=524
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 25/70 (35%), Positives = 34/70 (48%), Gaps = 14/70 (20%)
Query 13 LASGGADNRVRLWECDASKQWTELPALAG-EHHTDWVRDVSFRPQGASSFVVPGALLLAS 71
L + G D+ +R+W + K P +A EHHTDWV D+ G + L S
Sbjct 45 LFTAGRDSIIRIWSVNQHKD----PYIASMEHHTDWVNDIVLCCNGKT---------LIS 91
Query 72 CSEDKTVKLW 81
S D TVK+W
Sbjct 92 ASSDTTVKVW 101
Lambda K H
0.315 0.128 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40