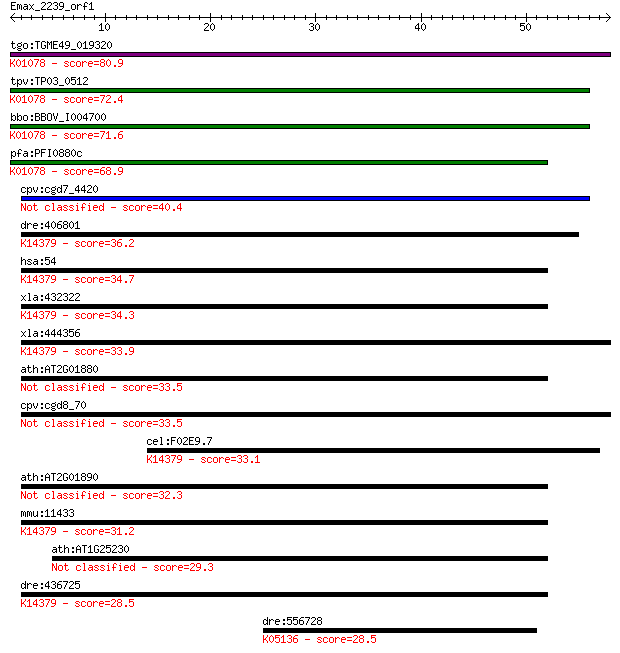
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2239_orf1
Length=57
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01... 80.9 1e-15
tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosp... 72.4 3e-13
bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01... 71.6 6e-13
pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3... 68.9 4e-12
cpv:cgd7_4420 secreted acid phosphatase (calcineurin family),s... 40.4 0.001
dre:406801 acp5a, acp5, sb:cb576, wu:fb19f01, wu:fb30b03, wu:f... 36.2 0.030
hsa:54 ACP5, MGC117378, TRAP; acid phosphatase 5, tartrate res... 34.7 0.081
xla:432322 acp5, MGC78938; acid phosphatase 5, tartrate resist... 34.3 0.098
xla:444356 MGC82831 protein; K14379 tartrate-resistant acid ph... 33.9 0.13
ath:AT2G01880 PAP7; PAP7 (PURPLE ACID PHOSPHATASE 7); acid pho... 33.5 0.18
cpv:cgd8_70 hypothetical protein 33.5 0.19
cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant ac... 33.1 0.20
ath:AT2G01890 PAP8; PAP8 (PURPLE ACID PHOSPHATASE 8); acid pho... 32.3 0.43
mmu:11433 Acp5, TRACP, TRAP; acid phosphatase 5, tartrate resi... 31.2 0.88
ath:AT1G25230 purple acid phosphatase family protein 29.3 3.3
dre:436725 acp5b, zgc:92339; acid phosphatase 5b, tartrate res... 28.5 5.8
dre:556728 crfb8, im:7156745, si:ch211-272f15.1, zgc:158705; c... 28.5 6.1
> tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=431
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/57 (64%), Positives = 43/57 (75%), Gaps = 0/57 (0%)
Query 1 PGSNFPNGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTAMRERTD 57
PGSNF GVS NDT+W FE+VYSD +G+LKMPF TVLG DDWS NYT+ RT+
Sbjct 89 PGSNFLGGVSSLNDTRWQSEFENVYSDANGALKMPFFTVLGVDDWSRNYTSEALRTE 145
> tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosphatase
[EC:3.1.3.2]
Length=404
Score = 72.4 bits (176), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 32/55 (58%), Positives = 41/55 (74%), Gaps = 0/55 (0%)
Query 1 PGSNFPNGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTAMRER 55
PG NF NGV+G ND KW + FESVY+D+SG + +P TVLG++DW G+Y A ER
Sbjct 68 PGFNFDNGVNGLNDEKWKKFFESVYNDDSGLMDLPMFTVLGSEDWLGDYNAQYER 122
> bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=395
Score = 71.6 bits (174), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 40/55 (72%), Gaps = 0/55 (0%)
Query 1 PGSNFPNGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTAMRER 55
PGSNF GV+G+NDTKWD F+SVY E GS+++P TVLGA DW G++ + R
Sbjct 66 PGSNFEYGVTGSNDTKWDTHFQSVYRSEDGSMEIPMFTVLGAGDWLGDFNSQINR 120
> pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3.2);
K01078 acid phosphatase [EC:3.1.3.2]
Length=396
Score = 68.9 bits (167), Expect = 4e-12, Method: Composition-based stats.
Identities = 30/51 (58%), Positives = 35/51 (68%), Gaps = 0/51 (0%)
Query 1 PGSNFPNGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTA 51
PGSNF +GV G ND W +E VYS+E G + MPF TVLG DW+GNY A
Sbjct 64 PGSNFIDGVKGLNDPAWKNLYEDVYSEEKGDMYMPFFTVLGTRDWTGNYNA 114
> cpv:cgd7_4420 secreted acid phosphatase (calcineurin family),signal
peptide
Length=826
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 25/55 (45%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Query 2 GSNFPN-GVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTAMRER 55
G NF GVS +D W+ FESV+ ES + F VLG DW GN TA +R
Sbjct 65 GDNFYRWGVSSVDDPIWENMFESVFDQESLQ-DVQFRCVLGNHDWWGNATAQVDR 118
> dre:406801 acp5a, acp5, sb:cb576, wu:fb19f01, wu:fb30b03, wu:fi14e01,
wu:fj66f03, zgc:63825; acid phosphatase 5a, tartrate
resistant (EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=339
Score = 36.2 bits (82), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 30/54 (55%), Gaps = 3/54 (5%)
Query 2 GSNFP-NGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTAMRE 54
G NF GV+ ND ++ E+FE VY+ + SL +P+ + G D GN A E
Sbjct 80 GDNFYYKGVTDVNDPRFQETFEDVYTQD--SLNIPWYVIAGNHDHVGNVKAQIE 131
> hsa:54 ACP5, MGC117378, TRAP; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=325
Score = 34.7 bits (78), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 29/51 (56%), Gaps = 2/51 (3%)
Query 2 GSNFP-NGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTA 51
G NF GV ND ++ E+FE V+SD S K+P+ + G D GN +A
Sbjct 70 GDNFYFTGVQDINDKRFQETFEDVFSDRSLR-KVPWYVLAGNHDHLGNVSA 119
> xla:432322 acp5, MGC78938; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=326
Score = 34.3 bits (77), Expect = 0.098, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 31/51 (60%), Gaps = 2/51 (3%)
Query 2 GSNFP-NGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTA 51
G NF +GV+ +D ++ +FESVYS ES +K P+ + G D GN +A
Sbjct 72 GDNFYYDGVTDVSDPRFKITFESVYSSES-LIKHPWYILAGNHDHKGNVSA 121
> xla:444356 MGC82831 protein; K14379 tartrate-resistant acid
phosphatase type 5 [EC:3.1.3.2]
Length=325
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 22/57 (38%), Positives = 34/57 (59%), Gaps = 2/57 (3%)
Query 2 GSNFP-NGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTAMRERTD 57
G NF +GV+ +D ++ +FESVYS ES +K+P+ + G D GN +A T+
Sbjct 71 GDNFYYDGVTDESDPRFKFTFESVYSAES-LVKLPWYILAGNHDHKGNVSAQIAYTN 126
> ath:AT2G01880 PAP7; PAP7 (PURPLE ACID PHOSPHATASE 7); acid phosphatase/
protein serine/threonine phosphatase
Length=328
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Query 2 GSNF-PNGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTA 51
G NF +G+ G ND ++ SF +Y+ SL+ + +VLG D+ GN A
Sbjct 80 GDNFYDDGLKGVNDPSFEASFSHIYTHP--SLQKQWYSVLGNHDYRGNVEA 128
> cpv:cgd8_70 hypothetical protein
Length=424
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 27/57 (47%), Gaps = 3/57 (5%)
Query 2 GSNF-PNGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTAMRERTD 57
G NF GV D W E E + S LK+ + LG DW G+ TA +RT+
Sbjct 69 GDNFYQRGVKSVEDAAWTEILEEPFGKLSKHLKVH--SCLGDHDWRGSTTAQIDRTN 123
> cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant
acid phosphatase type 5 [EC:3.1.3.2]
Length=419
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 28/43 (65%), Gaps = 2/43 (4%)
Query 14 DTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTAMRERT 56
D +++ FE+VY++ S L++P+LT+ G D GN TA E T
Sbjct 136 DPRFESRFENVYTNPS--LQVPWLTIAGNHDHFGNVTAEIEYT 176
> ath:AT2G01890 PAP8; PAP8 (PURPLE ACID PHOSPHATASE 8); acid phosphatase/
protein serine/threonine phosphatase
Length=335
Score = 32.3 bits (72), Expect = 0.43, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 31/51 (60%), Gaps = 3/51 (5%)
Query 2 GSNF-PNGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTA 51
G NF +G+ D+++ +SF ++Y+ + SL+ P+ VLG D+ GN A
Sbjct 84 GDNFYDDGIISPYDSQFQDSFTNIYT--ATSLQKPWYNVLGNHDYRGNVYA 132
> mmu:11433 Acp5, TRACP, TRAP; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=327
Score = 31.2 bits (69), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 2/51 (3%)
Query 2 GSNFP-NGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTA 51
G NF GV A+D ++ E+FE V+SD + +P+ + G D GN +A
Sbjct 72 GDNFYFTGVHDASDKRFQETFEDVFSDRALR-NIPWYVLAGNHDHLGNVSA 121
> ath:AT1G25230 purple acid phosphatase family protein
Length=339
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Query 5 FPNGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTA 51
+ NG+ +D + SF ++Y+ S SL+ P+ VLG D+ G+ A
Sbjct 89 YDNGMKSIDDPAFQLSFSNIYT--SPSLQKPWYLVLGNHDYRGDVEA 133
> dre:436725 acp5b, zgc:92339; acid phosphatase 5b, tartrate resistant;
K14379 tartrate-resistant acid phosphatase type 5
[EC:3.1.3.2]
Length=327
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 2 GSNFP-NGVSGANDTKWDESFESVYSDESGSLKMPFLTVLGADDWSGNYTA 51
G NF +GV +DT++ S+E V+S + +P+ + G D GN +A
Sbjct 73 GDNFYYDGVKDVDDTRFKFSYEQVFS-HPALMTIPWYLIAGNHDHRGNVSA 122
> dre:556728 crfb8, im:7156745, si:ch211-272f15.1, zgc:158705;
cytokine receptor family member b8; K05136 interleukin 20 receptor
alpha
Length=492
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 25 YSDESGSLKMPFLTVLGADDWSGNYT 50
+S ESG LK+P + +LG +D SG T
Sbjct 426 WSPESGELKIPLMGLLGLEDESGVQT 451
Lambda K H
0.308 0.128 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2082685716
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40