bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2276_orf1
Length=149
Score E
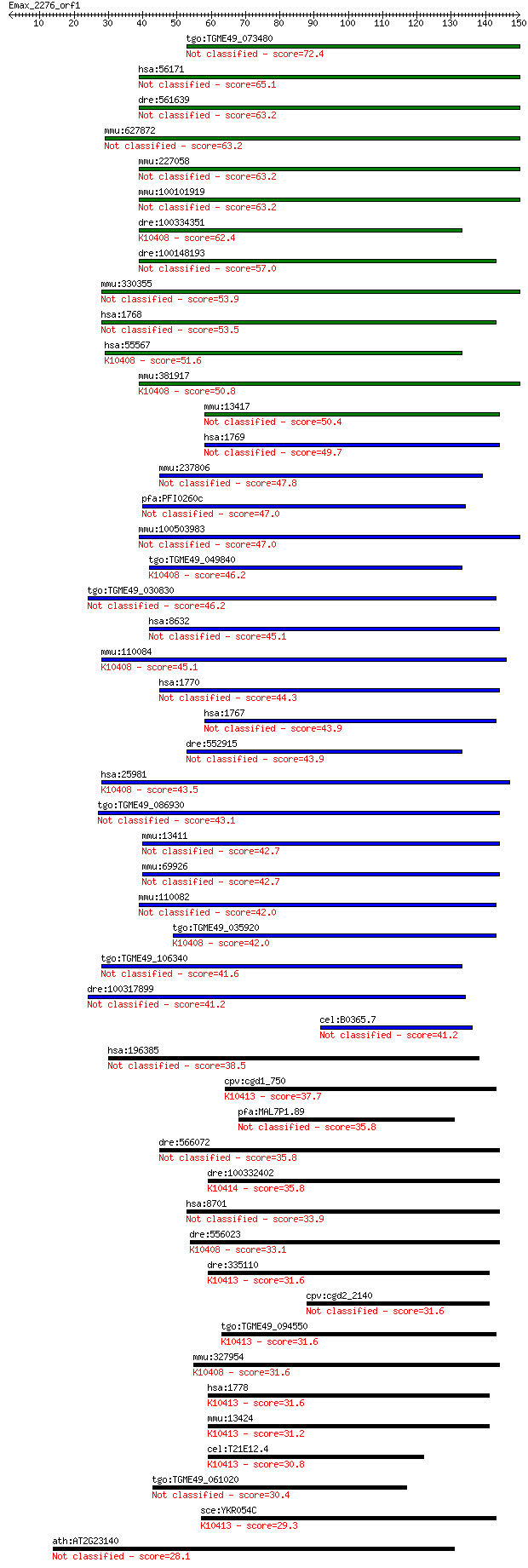
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_073480 axonemal beta dynein heavy chain, putative (... 72.4 5e-13
hsa:56171 DNAH7, DKFZp686C09101, FLJ37196, KIAA0944, MGC39580;... 65.1 9e-11
dre:561639 dnah7, si:ch211-284h20.3; dynein, axonemal, heavy c... 63.2 3e-10
mmu:627872 Dnahc7a, Dnahc7, Gm992; dynein, axonemal, heavy cha... 63.2 3e-10
mmu:227058 Dnahc7b, Gm107; dynein, axonemal, heavy chain 7B 63.2 3e-10
mmu:100101919 Dnahc7c; dynein, axonemal, heavy chain 7C 63.2 3e-10
dre:100334351 dnah3; dynein, axonemal, heavy chain 3; K10408 d... 62.4 6e-10
dre:100148193 dnah6; dynein, axonemal, heavy chain 6 57.0
mmu:330355 Dnahc6, 9830168K20, A730004I20Rik, KIAA1697, mKIAA1... 53.9 2e-07
hsa:1768 DNAH6, DKFZp667K087, DNHL1, Dnahc6, FLJ23529, FLJ3735... 53.5 2e-07
hsa:55567 DNAH3, DKFZp434N074, DLP3, DNAHC3B, FLJ31947, FLJ439... 51.6 1e-06
mmu:381917 Dnahc3, BC051401, Dnah3; dynein, axonemal, heavy ch... 50.8 2e-06
mmu:13417 Dnahc8, ATPase, Hst6.7b, P1-Loop; dynein, axonemal, ... 50.4 2e-06
hsa:1769 DNAH8, ATPase, FLJ25850, FLJ36115, FLJ36334, hdhc9; d... 49.7 3e-06
mmu:237806 Dnahc9, 9030022M04, A230091C01, C230051G16, D11Ertd... 47.8 1e-05
pfa:PFI0260c dynein heavy chain, putative 47.0 2e-05
mmu:100503983 dynein heavy chain 12, axonemal-like 47.0 2e-05
tgo:TGME49_049840 dynein heavy chain domain containing protein... 46.2 4e-05
tgo:TGME49_030830 dynein heavy chain, putative (EC:3.1.3.48) 46.2 4e-05
hsa:8632 DNAH17, DNAHL1, DNEL2, FLJ40457, MGC132767, MGC138489... 45.1 8e-05
mmu:110084 Dnahc1, B230373P09Rik, DKFZp434A236, Dnah1, E030034... 45.1 9e-05
hsa:1770 DNAH9, DNAH17L, DNEL1, DYH9, Dnahc9, HL-20, HL20, KIA... 44.3 1e-04
hsa:1767 DNAH5, CILD3, DNAHC5, FLJ46759, HL1, KIAA1603, KTGNR,... 43.9 2e-04
dre:552915 dnah9l, dhc9; dynein, axonemal, heavy polypeptide 9... 43.9 2e-04
hsa:25981 DNAH1, DNAHC1, HDHC7, HL-11, HL11, XLHSRF-1; dynein,... 43.5 3e-04
tgo:TGME49_086930 dynein heavy chain, putative (EC:2.7.4.14) 43.1 3e-04
mmu:13411 Dnahc11, iv, lrd; dynein, axonemal, heavy chain 11 42.7
mmu:69926 Dnahc17, 2810003K23Rik, Dnah17, Dnahcl1, Gm1178, mKI... 42.7 4e-04
mmu:110082 Dnahc5, AU022615, Dnah5, KIAA1603, Mdnah5, mKIAA160... 42.0 7e-04
tgo:TGME49_035920 dynein 1-beta heavy chain, flagellar inner a... 42.0 8e-04
tgo:TGME49_106340 dynein gamma chain, flagellar outer arm, put... 41.6 9e-04
dre:100317899 dnah1; dynein, axonemal, heavy chain 1 41.2
cel:B0365.7 dynein; hypothetical protein 41.2 0.001
hsa:196385 DNAH10, FLJ38262, FLJ43486, FLJ43808, KIAA2017; dyn... 38.5 0.007
cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain 1,... 37.7 0.015
pfa:MAL7P1.89 dynein heavy chain, putative 35.8 0.048
dre:566072 dynein, axonemal, heavy polypeptide 11-like 35.8 0.049
dre:100332402 cytoplasmic dynein 2 heavy chain 1-like; K10414 ... 35.8 0.053
hsa:8701 DNAH11, CILD7, DNAHBL, DNAHC11, DNHBL, DPL11, FLJ3009... 33.9 0.19
dre:556023 dynein, axonemal, heavy chain 2-like; K10408 dynein... 33.1 0.32
dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic 1... 31.6 0.96
cpv:cgd2_2140 hypothetical protein 31.6 0.99
tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein... 31.6 1.0
mmu:327954 Dnahc2, 2900022L05Rik, 4930564A01, D130094J20, D330... 31.6 1.0
hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNE... 31.6 1.0
mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,... 31.2 1.1
cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1); ... 30.8 1.6
tgo:TGME49_061020 axonemal dynein heavy chain, putative (EC:2.... 30.4 2.4
sce:YKR054C DYN1, DHC1, PAC6; Cytoplasmic heavy chain dynein, ... 29.3 4.5
ath:AT2G23140 binding / ubiquitin-protein ligase 28.1 9.7
> tgo:TGME49_073480 axonemal beta dynein heavy chain, putative
(EC:5.99.1.3)
Length=4273
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 61/97 (62%), Gaps = 0/97 (0%)
Query 53 YRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQ 112
YR E RR +++TP S++ FL F ++ + + + + + QY +GL+K+H S QV +Q+
Sbjct 2750 YRQELRRHYYITPRSYIEFLKLFSHLSVAKREELNRQVDQYRVGLEKIHSTSTQVGKIQE 2809
Query 113 QLEKLQPELAKSSMETQQLMQVLTVKQEHAATTMSLV 149
+LE L+P+L K+S + LM ++ QE AA T + V
Sbjct 2810 ELETLKPQLIKASQDMVDLMVDISKMQESAARTKATV 2846
> hsa:56171 DNAH7, DKFZp686C09101, FLJ37196, KIAA0944, MGC39580;
dynein, axonemal, heavy chain 7
Length=4024
Score = 65.1 bits (157), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 61/111 (54%), Gaps = 0/111 (0%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
C ST +++ + VE +R+ +VTP S+L + F + + S+ +++YE+GL+
Sbjct 2534 CKSFHTSTIDLSKSFFVELQRYNYVTPTSYLELISTFKLLLEKKRSEVMKMKKRYEVGLE 2593
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATTMSLV 149
KL S QV MQ +LE L P+L +S E ++M ++ + A T +V
Sbjct 2594 KLDSASSQVATMQMELEALHPQLKVASKEVDEMMIMIEKESVEVAKTEKIV 2644
> dre:561639 dnah7, si:ch211-284h20.3; dynein, axonemal, heavy
chain 7
Length=3990
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 63/111 (56%), Gaps = 0/111 (0%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
C ST +++ + E +R+ +VTP S+L + F ++ + ++ + +YE+GLQ
Sbjct 2500 CKSFHTSTIDLSAHFLSELQRYNYVTPTSYLELISMFKHLLQRKRTEVMKLKSRYEVGLQ 2559
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATTMSLV 149
KL + QV MQ +LE LQP+L ++ E +++M V+ + E + T +V
Sbjct 2560 KLESAATQVSTMQGELEALQPQLRVATKEVEEMMVVIQHESEEVSKTEKVV 2610
> mmu:627872 Dnahc7a, Dnahc7, Gm992; dynein, axonemal, heavy chain
7A
Length=4068
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 36/122 (29%), Positives = 64/122 (52%), Gaps = 1/122 (0%)
Query 29 NENLKHLC-GACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHH 87
+E ++ C C ST N++ + E +R+ +VTP S+L + F + + ++
Sbjct 2567 SEEIREGCIDMCKRFHTSTINLSTSFHNELQRYNYVTPTSYLELISTFKLLLEKKRNEVM 2626
Query 88 HKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATTMS 147
+++YE+GL KL S QV MQ +LE L P+L +S E ++M ++ + A T
Sbjct 2627 KMKRRYEVGLDKLDSASSQVATMQSELEALHPQLKVASREVDEMMIIIERESMEVAKTEK 2686
Query 148 LV 149
+V
Sbjct 2687 IV 2688
> mmu:227058 Dnahc7b, Gm107; dynein, axonemal, heavy chain 7B
Length=4068
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 59/111 (53%), Gaps = 0/111 (0%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
C ST N++ + E +R+ +VTP S+L + F + + ++ +++YE+GL
Sbjct 2578 CKRFHTSTINLSTSFHNELQRYNYVTPTSYLELISTFKLLLEKKRNEVMKMKRRYEVGLD 2637
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATTMSLV 149
KL S QV MQ +LE L P+L +S E ++M ++ + A T +V
Sbjct 2638 KLDSASSQVATMQSELEALHPQLKVASREVDEMMIIIERESMEVAKTEKIV 2688
> mmu:100101919 Dnahc7c; dynein, axonemal, heavy chain 7C
Length=4092
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 59/111 (53%), Gaps = 0/111 (0%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
C ST N++ + E +R+ +VTP S+L + F + + ++ +++YE+GL
Sbjct 2602 CKRFHTSTINLSTSFHNELQRYNYVTPTSYLELISTFKLLLEKKRNEVMKMKRRYEVGLD 2661
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATTMSLV 149
KL S QV MQ +LE L P+L +S E ++M ++ + A T +V
Sbjct 2662 KLDSASSQVATMQSELEALHPQLKVASREVDEMMIIIERESMEVAKTEKIV 2712
> dre:100334351 dnah3; dynein, axonemal, heavy chain 3; K10408
dynein heavy chain, axonemal
Length=3972
Score = 62.4 bits (150), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 34/94 (36%), Positives = 56/94 (59%), Gaps = 0/94 (0%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
C ES + +++ Y + RR +VTP S+L + F + ++ S+ + + +Y +GLQ
Sbjct 2480 CKTFQESVRELSERYYSQLRRHNYVTPTSYLELILTFKALLNSKRSEVNELRNRYIVGLQ 2539
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLM 132
KL S QV MQQ+L LQPEL +++ +T Q+M
Sbjct 2540 KLDFASSQVAVMQQELTALQPELIETAKQTDQMM 2573
> dre:100148193 dnah6; dynein, axonemal, heavy chain 6
Length=4163
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/104 (30%), Positives = 57/104 (54%), Gaps = 0/104 (0%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
C +I S ++A+ + E RR ++ TP S+L ++ + + + Q + + + GL
Sbjct 2642 CVEIHVSVTDMAERFYSELRRHYYTTPTSYLELINLYLGMLGEKRQQLQAARDRIKNGLT 2701
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHA 142
KL + + V M+Q L L+P LA+ S++ LM+ L V QE+A
Sbjct 2702 KLLETNELVDKMKQDLSALEPVLAQKSIDVSALMEKLAVDQENA 2745
> mmu:330355 Dnahc6, 9830168K20, A730004I20Rik, KIAA1697, mKIAA1697,
mdhc6; dynein, axonemal, heavy chain 6
Length=4144
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 64/123 (52%), Gaps = 1/123 (0%)
Query 28 SNENLKH-LCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQH 86
N++L+ L C ++ S ++A+ Y E RR ++ TP S+L ++ + + + Q
Sbjct 2621 GNDDLREKLSLMCVNVHLSVSHMAERYYNELRRRYYTTPTSYLELINLYLTMLTEKRKQL 2680
Query 87 HHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATTM 146
+ + + GL KL + ++ V M+ L L+P L + S + + LM+ L V QE A
Sbjct 2681 VSARDRVKNGLTKLLETNVLVDKMKLDLSALEPVLLQKSQDVEALMEKLVVDQESADQVR 2740
Query 147 SLV 149
++V
Sbjct 2741 NVV 2743
> hsa:1768 DNAH6, DKFZp667K087, DNHL1, Dnahc6, FLJ23529, FLJ37357,
HL-2, HL2, KIAA1697, MGC132544, MGC141984, MGC177197; dynein,
axonemal, heavy chain 6
Length=4158
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 35/116 (30%), Positives = 59/116 (50%), Gaps = 1/116 (0%)
Query 28 SNENLKH-LCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQH 86
NE LK L C ++ S ++A+ Y E RR ++ TP S+L ++ + + + Q
Sbjct 2626 GNEELKEKLPLMCVNVHLSVSSMAERYYNELRRRYYTTPTSYLELINLYLSMLSEKRKQI 2685
Query 87 HHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHA 142
+ + + GL KL + ++ V M+ L L+P L S + + LM+ L V QE A
Sbjct 2686 ISARDRVKNGLTKLLETNILVDKMKLDLSALEPVLLAKSEDVEALMEKLAVDQESA 2741
> hsa:55567 DNAH3, DKFZp434N074, DLP3, DNAHC3B, FLJ31947, FLJ43919,
FLJ43964, Hsadhc3; dynein, axonemal, heavy chain 3; K10408
dynein heavy chain, axonemal
Length=4116
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 33/105 (31%), Positives = 57/105 (54%), Gaps = 1/105 (0%)
Query 29 NENLK-HLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHH 87
++N++ + C ES K ++ Y + RR +VTP S+L + F + ++ +
Sbjct 2614 DDNIRVEVVSMCKYFQESVKKLSLDYYNKLRRHNYVTPTSYLELILTFKTLLNSKRQEVA 2673
Query 88 HKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLM 132
+ +Y GLQKL + QV MQ++L LQP+L +S ET ++M
Sbjct 2674 MMRNRYLTGLQKLDFAASQVAVMQRELTALQPQLILTSEETAKMM 2718
> mmu:381917 Dnahc3, BC051401, Dnah3; dynein, axonemal, heavy
chain 3; K10408 dynein heavy chain, axonemal
Length=4088
Score = 50.8 bits (120), Expect = 2e-06, Method: Composition-based stats.
Identities = 34/111 (30%), Positives = 55/111 (49%), Gaps = 0/111 (0%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
C ES K ++ Y R +VTP S+L + F + ++ + + +Y GLQ
Sbjct 2597 CKYFQESVKKLSVDYYNTLLRHNYVTPTSYLELILTFKTLLNSKRQEVDTIRNRYLAGLQ 2656
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATTMSLV 149
KL S QV MQ +L LQP+L ++S +T +M + ++ + A LV
Sbjct 2657 KLEFASSQVAVMQVELTALQPQLIQTSEDTAMMMVKIELETKEADAKKLLV 2707
> mmu:13417 Dnahc8, ATPase, Hst6.7b, P1-Loop; dynein, axonemal,
heavy chain 8
Length=4731
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 49/86 (56%), Gaps = 0/86 (0%)
Query 58 RRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKL 117
RR HVTP S+L F++G+ ++ + + + ++ +GL KL + S V + Q L
Sbjct 3273 RRRAHVTPKSYLSFINGYKSIYTDKVKYINEQAERMNIGLDKLMEASESVAKLSQDLAVK 3332
Query 118 QPELAKSSMETQQLMQVLTVKQEHAA 143
+ ELA +S++ +++ +TV + +A
Sbjct 3333 EKELAVASIKADEVLAEVTVSAQASA 3358
> hsa:1769 DNAH8, ATPase, FLJ25850, FLJ36115, FLJ36334, hdhc9;
dynein, axonemal, heavy chain 8
Length=4490
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 49/86 (56%), Gaps = 0/86 (0%)
Query 58 RRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKL 117
RR HVTP S+L F++G+ ++ + + + ++ +GL KL + S V + Q L
Sbjct 3032 RRRAHVTPKSYLSFINGYKNIYAEKVKFINEQAERMNIGLDKLMEASESVAKLSQDLAVK 3091
Query 118 QPELAKSSMETQQLMQVLTVKQEHAA 143
+ ELA +S++ +++ +TV + +A
Sbjct 3092 EKELAVASIKADEVLAEVTVSAQASA 3117
> mmu:237806 Dnahc9, 9030022M04, A230091C01, C230051G16, D11Ertd686e,
DNAH9, KIAA0357, mKIAA0357; dynein, axonemal, heavy
chain 9
Length=4484
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 49/94 (52%), Gaps = 0/94 (0%)
Query 45 STKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVS 104
S + Y ++R+ + TP SFL F+ + + + K ++ E GL KLH S
Sbjct 3019 SVNKTSQSYLTNEQRYNYTTPKSFLEFIRLYQSLLERNGKELQAKVERLENGLLKLHSTS 3078
Query 105 MQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVK 138
QV D++ +L + EL + +T +L+QV+ V+
Sbjct 3079 AQVDDLKAKLATQEVELRHKNEDTDKLIQVVGVE 3112
> pfa:PFI0260c dynein heavy chain, putative
Length=6118
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 23/94 (24%), Positives = 51/94 (54%), Gaps = 0/94 (0%)
Query 40 ADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQK 99
A + + D Y+ RR +VTP S+L F+D + +++ + + ++ ++GL+K
Sbjct 4321 AIVHNKVSDTCDTYKERMRRNTYVTPKSYLSFIDLYKQMYVKKYDEIKCLKESVDIGLKK 4380
Query 100 LHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQ 133
L++ +M V M++ L + +L +S + L++
Sbjct 4381 LNEAAMDVQKMRESLTSEEEKLKESDEQMNILLE 4414
> mmu:100503983 dynein heavy chain 12, axonemal-like
Length=2998
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/111 (24%), Positives = 52/111 (46%), Gaps = 0/111 (0%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
C S ++++ + E R +VT S+L + F + + +Q+Y GL
Sbjct 1506 CKHFHTSIMHLSERFLEELGRHNYVTATSYLELIGSFRQLLTKKRQAVMEAKQRYVNGLD 1565
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATTMSLV 149
+L QV +M+ +L +LQP+L + +E ++MQ++ V+ V
Sbjct 1566 QLAFAESQVGEMKLELVELQPKLEAAKIENARMMQIIEVESAQVEAKRKFV 1616
> tgo:TGME49_049840 dynein heavy chain domain containing protein
; K10408 dynein heavy chain, axonemal
Length=4140
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 46/91 (50%), Gaps = 0/91 (0%)
Query 42 IFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLH 101
I S ++ A+ + E RR VTP FL L ++ + ++++ GL KL
Sbjct 2651 IHRSVQDAAEKFLAECRRPVFVTPTCFLELLKTLTETVKSKQLELSTIRERFGKGLGKLA 2710
Query 102 DVSMQVMDMQQQLEKLQPELAKSSMETQQLM 132
+ + QV MQQQL++ P L +S E +Q M
Sbjct 2711 EAARQVERMQQQLQEWHPVLVATSEEVEQKM 2741
> tgo:TGME49_030830 dynein heavy chain, putative (EC:3.1.3.48)
Length=4713
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 53/119 (44%), Gaps = 0/119 (0%)
Query 24 PSADSNENLKHLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRA 83
P + +N+ L AD + E R + T +SF+ FL + ++ T
Sbjct 3323 PDLIAPDNISTLSAVSADFHAYAVAAQAAFYEETGRRVYTTGSSFVDFLQCYLHLLDTER 3382
Query 84 SQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHA 142
+ + + Y G+ +L + V ++ L K QP L KS+ +TQQLM L ++ A
Sbjct 3383 AAIQERTRHYRRGISRLEATTKLVEQLRVDLLKSQPVLEKSTRDTQQLMASLERDRQRA 3441
> hsa:8632 DNAH17, DNAHL1, DNEL2, FLJ40457, MGC132767, MGC138489;
dynein, axonemal, heavy chain 17
Length=4462
Score = 45.1 bits (105), Expect = 8e-05, Method: Composition-based stats.
Identities = 24/102 (23%), Positives = 52/102 (50%), Gaps = 0/102 (0%)
Query 42 IFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLH 101
+ + ++ Y +RR+ + TP +FL + + + + ++ K ++ E GL KL
Sbjct 2994 VHTTVNEMSRVYLATERRYNYTTPKTFLEQIKLYQNLLAKKRTELVAKIERLENGLMKLQ 3053
Query 102 DVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAA 143
+ QV D++ +L + EL + + QL+QV+ ++ E +
Sbjct 3054 STASQVDDLKAKLAIQEAELKQKNESADQLIQVVGIEAEKVS 3095
> mmu:110084 Dnahc1, B230373P09Rik, DKFZp434A236, Dnah1, E030034C22Rik,
MDHC7, MGC37121; dynein, axonemal, heavy chain 1;
K10408 dynein heavy chain, axonemal
Length=4250
Score = 45.1 bits (105), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 56/118 (47%), Gaps = 0/118 (0%)
Query 28 SNENLKHLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHH 87
S E ++ L C I +S + Y E R +VTP S+L L+ F + + +
Sbjct 2752 SEEVIQGLIQVCVFIHQSVASKCVEYLAELARHNYVTPKSYLELLNIFSILIGQKKMELK 2811
Query 88 HKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAATT 145
+ + + GL KL S V MQ++LE ++P L +++ +T M+ + V A T
Sbjct 2812 TAKNRMKSGLDKLLRTSEDVAKMQEELEIMRPLLEEAAKDTMLTMEQIKVDTAIAEET 2869
> hsa:1770 DNAH9, DNAH17L, DNEL1, DYH9, Dnahc9, HL-20, HL20, KIAA0357;
dynein, axonemal, heavy chain 9
Length=4486
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 51/99 (51%), Gaps = 0/99 (0%)
Query 45 STKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVS 104
S + Y ++R+ + TP SFL F+ + + + K ++ E GL KLH S
Sbjct 3021 SVNQTSQSYLSNEQRYNYTTPKSFLEFIRLYQSLLHRHRKELKCKTERLENGLLKLHSTS 3080
Query 105 MQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAA 143
QV D++ +L + EL + + + +L+QV+ V+ + +
Sbjct 3081 AQVDDLKAKLAAQEVELKQKNEDADKLIQVVGVETDKVS 3119
> hsa:1767 DNAH5, CILD3, DNAHC5, FLJ46759, HL1, KIAA1603, KTGNR,
PCD; dynein, axonemal, heavy chain 5
Length=4624
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 47/85 (55%), Gaps = 0/85 (0%)
Query 58 RRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKL 117
RR HVTP S+L F+ G+ +++ + + + GL+KL + S V + ++LE
Sbjct 3168 RRSTHVTPKSYLSFIQGYKFIYGEKHVEVRTLANRMNTGLEKLKEASESVAALSKELEAK 3227
Query 118 QPELAKSSMETQQLMQVLTVKQEHA 142
+ EL ++ + +++ +T+K + A
Sbjct 3228 EKELQVANDKADMVLKEVTMKAQAA 3252
> dre:552915 dnah9l, dhc9; dynein, axonemal, heavy polypeptide
9 like
Length=4478
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/80 (30%), Positives = 46/80 (57%), Gaps = 0/80 (0%)
Query 53 YRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQ 112
Y+ ++RF + TP SFL F+ + + ++ ++ K ++ E GLQKL + QV D++
Sbjct 3020 YQQNEKRFNYTTPKSFLEFMKLYGNLLGSKRTELRQKTERLENGLQKLLTTASQVEDLKA 3079
Query 113 QLEKLQPELAKSSMETQQLM 132
+L + EL + +T+ L+
Sbjct 3080 KLAIQEVELHLRNTDTEALI 3099
> hsa:25981 DNAH1, DNAHC1, HDHC7, HL-11, HL11, XLHSRF-1; dynein,
axonemal, heavy chain 1; K10408 dynein heavy chain, axonemal
Length=4265
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 54/122 (44%), Gaps = 3/122 (2%)
Query 28 SNENLKHLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHH 87
S E ++ L C I +S Y E R +VTP S+L L F + + +
Sbjct 2767 SQEEIQGLIQVCVYIHQSVSKKCIEYLAELTRHNYVTPKSYLELLHIFSILIGQKKLELK 2826
Query 88 HKQQQYELGLQKLHDVSMQVMDMQQQLEKLQP---ELAKSSMETQQLMQVLTVKQEHAAT 144
+ + + GL KL S V MQ+ LE + P E AK +M T + ++V T E
Sbjct 2827 TAKNRMKSGLDKLLRTSEDVAKMQEDLESMHPLLEEAAKDTMLTMEQIKVDTAIAEETRN 2886
Query 145 TM 146
++
Sbjct 2887 SV 2888
> tgo:TGME49_086930 dynein heavy chain, putative (EC:2.7.4.14)
Length=4720
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 54/117 (46%), Gaps = 2/117 (1%)
Query 27 DSNENLKHLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQH 86
D N + C + +S + + + R +VTP FL + + R
Sbjct 3138 DVEANAQRRAYLCVQMHQSAIETSKAFLTQLNRPVYVTPKCFLDLIGLVVKLKNERQDAL 3197
Query 87 HHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAA 143
++ GL +LH + QV ++++LE+L+P L +E++QL+ TV+ + AA
Sbjct 3198 QRQRSLLSAGLTRLHRTNAQVEKLREELEQLKPVLETKKIESEQLLA--TVESDRAA 3252
> mmu:13411 Dnahc11, iv, lrd; dynein, axonemal, heavy chain 11
Length=4488
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 25/104 (24%), Positives = 53/104 (50%), Gaps = 0/104 (0%)
Query 40 ADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQK 99
A + S K ++ Y +RR+ + TP SFL+ + F + + + KQ+ G+QK
Sbjct 3018 AHVHTSVKEVSAWYYQNERRYNYTTPRSFLKQISLFKSLLKKKREEVKQKQEHLGNGIQK 3077
Query 100 LHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAA 143
L + QV +++ +L + EL +++ + L+ + ++ E +
Sbjct 3078 LQTTASQVGNLKSRLASQEAELQLRNLDAEALITKIGLQTEKVS 3121
> mmu:69926 Dnahc17, 2810003K23Rik, Dnah17, Dnahcl1, Gm1178, mKIAA3028;
dynein, axonemal, heavy chain 17
Length=4453
Score = 42.7 bits (99), Expect = 4e-04, Method: Composition-based stats.
Identities = 25/104 (24%), Positives = 52/104 (50%), Gaps = 0/104 (0%)
Query 40 ADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQK 99
A + + ++ Y +RR+ + TP +FL + + + + + K ++ E GL K
Sbjct 2976 AYVHTTVNEMSKIYLTIERRYNYTTPKTFLEQIKLYQNLLAKKRMELVAKIERLENGLMK 3035
Query 100 LHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAA 143
L + QV D++ +L + EL + + +L+QV+ V+ E +
Sbjct 3036 LQSTASQVDDLKAKLAVQETELKQKNENADKLIQVVGVETEKVS 3079
> mmu:110082 Dnahc5, AU022615, Dnah5, KIAA1603, Mdnah5, mKIAA1603;
dynein, axonemal, heavy chain 5
Length=4621
Score = 42.0 bits (97), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 26/104 (25%), Positives = 51/104 (49%), Gaps = 11/104 (10%)
Query 39 CADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQ 98
CAD F+ RR HVTP S+L F+ G+ +++ + + + GL+
Sbjct 3157 CADYFQRF-----------RRSTHVTPKSYLSFIQGYKFIYEEKHMEVQSLANRMNTGLE 3205
Query 99 KLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHA 142
KL + S V + ++L + EL ++ + +++ +T+K + A
Sbjct 3206 KLKEASESVAALSKELAGKEKELQVANEKADTVLKEVTMKAQAA 3249
> tgo:TGME49_035920 dynein 1-beta heavy chain, flagellar inner
arm I1 complex, putative ; K10408 dynein heavy chain, axonemal
Length=4213
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 28/96 (29%), Positives = 48/96 (50%), Gaps = 2/96 (2%)
Query 49 IADCYRV--EQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQ 106
IA+ R+ E +R +VTP FL + G+ ++ + + + +GL KL + Q
Sbjct 2743 IAESERMFAEMKRRNYVTPTKFLELVQGYIRLYREKTEEVQELVHKLTVGLHKLVETRAQ 2802
Query 107 VMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHA 142
V M +LE+ + +AK E Q L+ V+ K+ A
Sbjct 2803 VEVMGTELERKKEIVAKKQTECQDLLVVIVEKRSVA 2838
> tgo:TGME49_106340 dynein gamma chain, flagellar outer arm, putative
Length=4157
Score = 41.6 bits (96), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 27/105 (25%), Positives = 51/105 (48%), Gaps = 0/105 (0%)
Query 28 SNENLKHLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHH 87
S E L A I + + Y + RR +VTP S+L FL + V+ + + +
Sbjct 2606 SPEKKATLFSAMGRIHTDVGRMCEEYFLRMRRHVYVTPKSYLSFLSFYKLVYAEKFKEVN 2665
Query 88 HKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLM 132
+ + +GL KL+ + + M+ +L+ + +L +S +T QL+
Sbjct 2666 NLEHSVNVGLLKLNQAAQDIKQMKVKLKDEEKKLRESEEQTNQLL 2710
> dre:100317899 dnah1; dynein, axonemal, heavy chain 1
Length=1874
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/117 (27%), Positives = 58/117 (49%), Gaps = 7/117 (5%)
Query 24 PSADSN-ENLKHLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTR 82
P +SN + ++ L C +I + + Y E R VTP S+L L F + +
Sbjct 365 PELESNPKTIQSLALMCVEIHQMVSRKCEQYLAELSRHNSVTPKSYLELLSIFSSLIGQK 424
Query 83 ASQHHHKQQQYELGLQK--LHDVSM----QVMDMQQQLEKLQPELAKSSMETQQLMQ 133
+ H +Q+ + GL K L + S+ V MQ++LE ++P+L +++ +T M+
Sbjct 425 KQELHSARQRMKTGLDKVILLNFSLVXXEDVAKMQEELEMMRPQLEEAAKDTVITME 481
> cel:B0365.7 dynein; hypothetical protein
Length=2769
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 29/44 (65%), Gaps = 0/44 (0%)
Query 92 QYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVL 135
+YE G++K+ QV MQ +L +LQP+L ++S+ET LM +
Sbjct 2124 KYEKGMEKMKRAEEQVAGMQGELLRLQPQLVRTSIETSMLMSTI 2167
> hsa:196385 DNAH10, FLJ38262, FLJ43486, FLJ43808, KIAA2017; dynein,
axonemal, heavy chain 10
Length=4471
Score = 38.5 bits (88), Expect = 0.007, Method: Composition-based stats.
Identities = 22/108 (20%), Positives = 59/108 (54%), Gaps = 0/108 (0%)
Query 30 ENLKHLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHK 89
EN++++ + +S + + + + RR +VTP ++L F++ + + + + +
Sbjct 2984 ENIENVVKHVVLVHQSVDHYSQQFLQKLRRSNYVTPKNYLDFINTYSKLLDEKTQCNIAQ 3043
Query 90 QQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTV 137
++ + GL KL + ++Q+ ++ Q+L + + LA+ S + L++ + V
Sbjct 3044 CKRLDGGLDKLKEATIQLDELNQKLAEQKIVLAEKSAACEALLEEIAV 3091
> cpv:cgd1_750 dynein heavy chain ; K10413 dynein heavy chain
1, cytosolic
Length=5246
Score = 37.7 bits (86), Expect = 0.015, Method: Composition-based stats.
Identities = 21/79 (26%), Positives = 36/79 (45%), Gaps = 0/79 (0%)
Query 64 TPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAK 123
TP F FL ++ + +QQ GL+ L +V +QQ+L + + L
Sbjct 3553 TPRDFFDFLKHIIKIYKEKNETLLEQQQHLSSGLETLRSTEQEVATLQQELGEKEKILIA 3612
Query 124 SSMETQQLMQVLTVKQEHA 142
++E +Q MQ + +Q A
Sbjct 3613 KNVEAEQKMQQMIKEQGEA 3631
> pfa:MAL7P1.89 dynein heavy chain, putative
Length=5846
Score = 35.8 bits (81), Expect = 0.048, Method: Composition-based stats.
Identities = 17/63 (26%), Positives = 31/63 (49%), Gaps = 0/63 (0%)
Query 68 FLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELAKSSME 127
+L FL+ F Y+F + ++ Y L KLH + M+ L +QP L +++E
Sbjct 3957 YLHFLEHFDYLFFIKKKEYDKHIDLYSKALHKLHQCEEDIKIMKNNLLNIQPILNSTNIE 4016
Query 128 TQQ 130
++
Sbjct 4017 MKK 4019
> dre:566072 dynein, axonemal, heavy polypeptide 11-like
Length=1584
Score = 35.8 bits (81), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 47/99 (47%), Gaps = 0/99 (0%)
Query 45 STKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVS 104
S K +++ Y ++R + T SFL L + + R + H + + + GLQK+ +
Sbjct 118 SVKRVSERYCCNEKRHNYSTSKSFLEQLRLYKSLLENREKELHQRLDRLQSGLQKIKSTA 177
Query 105 MQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQEHAA 143
QV D+ +L + EL + + LM + ++ E +
Sbjct 178 SQVEDLTVKLNSQEVELTMKNQAAEALMARIGLQTERVS 216
> dre:100332402 cytoplasmic dynein 2 heavy chain 1-like; K10414
dynein heavy chain 2, cytosolic
Length=3301
Score = 35.8 bits (81), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 20/85 (23%), Positives = 43/85 (50%), Gaps = 0/85 (0%)
Query 59 RFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQ 118
R F TP+ +L FL + ++ ++ + HKQQ + G+ KL++ V +++++ +
Sbjct 2735 REFGATPSQYLNFLQVYQSIYSSKRKELTHKQQHLQAGVAKLNEAKALVDELKRRAAEQS 2794
Query 119 PELAKSSMETQQLMQVLTVKQEHAA 143
L E +Q +T ++A+
Sbjct 2795 SLLKTKQAEADAALQEITTSMQNAS 2819
> hsa:8701 DNAH11, CILD7, DNAHBL, DNAHC11, DNHBL, DPL11, FLJ30095,
FLJ37699; dynein, axonemal, heavy chain 11
Length=4523
Score = 33.9 bits (76), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 22/91 (24%), Positives = 46/91 (50%), Gaps = 0/91 (0%)
Query 53 YRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQ 112
Y +RR + TP SFL + F + + ++ K+++ G+QKL + QV D++
Sbjct 3066 YYQNERRHNYTTPKSFLEQISLFKNLLKKKQNEVSEKKERLVNGIQKLKTTASQVGDLKA 3125
Query 113 QLEKLQPELAKSSMETQQLMQVLTVKQEHAA 143
+L + EL + + + L+ + ++ E +
Sbjct 3126 RLASQEAELQLRNHDAEALITKIGLQTEKVS 3156
> dre:556023 dynein, axonemal, heavy chain 2-like; K10408 dynein
heavy chain, axonemal
Length=4424
Score = 33.1 bits (74), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 21/90 (23%), Positives = 48/90 (53%), Gaps = 1/90 (1%)
Query 54 RVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQ 113
+VE +R +VTP ++L + G+ + + + + + GL K+ D +V M +
Sbjct 2963 KVELKRHNYVTPTNYLELVSGYKKLLSEKRDELGEQVSKLRNGLFKIDDTRTKVEAMSVE 3022
Query 114 LEKLQPELAKSSMETQQLMQVLTVKQEHAA 143
LE+ + ++A+ + ++ + V+ V+Q+ A
Sbjct 3023 LEEAKKKVAEFQKQCEEYL-VVIVQQKREA 3051
> dre:335110 dync1h1, fk70a07, wu:fk70a07; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4643
Score = 31.6 bits (70), Expect = 0.96, Method: Compositional matrix adjust.
Identities = 20/83 (24%), Positives = 44/83 (53%), Gaps = 1/83 (1%)
Query 59 RFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQ 118
R +TP +L F++ + +F + S+ +Q +GL+K+ + QV ++++ L
Sbjct 3166 RTMAITPRHYLDFINHYANLFNEKRSELEEQQMHLNVGLRKIKETVDQVEELRRDLRIKS 3225
Query 119 PEL-AKSSMETQQLMQVLTVKQE 140
EL K++ +L +++ +QE
Sbjct 3226 QELEVKNAAANDKLKKMVKDQQE 3248
> cpv:cgd2_2140 hypothetical protein
Length=1818
Score = 31.6 bits (70), Expect = 0.99, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 36/57 (63%), Gaps = 7/57 (12%)
Query 88 HKQQQYELGLQKLHD----VSMQVMDMQQQLEKLQPELAKSSMETQQLMQVLTVKQE 140
+K QQ G+Q++ D +S+Q++ +Q+ L K++ K SM Q+LM++ VK++
Sbjct 1621 NKDQQ---GIQEIRDYLGNISLQLLLIQEHLVKIEFLRCKLSMIRQELMKIEDVKKD 1674
> tgo:TGME49_094550 dynein heavy chain, putative ; K10413 dynein
heavy chain 1, cytosolic
Length=4937
Score = 31.6 bits (70), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 38/80 (47%), Gaps = 0/80 (0%)
Query 63 VTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQPELA 122
+TP FL FL F + +A +Q+ + GLQ L QV +M+ L + + L
Sbjct 3320 MTPRDFLDFLHHFVNLVGEKADATGEQQRHLQAGLQTLRVAEEQVAEMRSALTEKESVLT 3379
Query 123 KSSMETQQLMQVLTVKQEHA 142
+ + E ++ M + +Q A
Sbjct 3380 EKNEEAEKKMGQMVEQQAEA 3399
> mmu:327954 Dnahc2, 2900022L05Rik, 4930564A01, D130094J20, D330014H01Rik,
Dnah2, Dnhd3; dynein, axonemal, heavy chain 2;
K10408 dynein heavy chain, axonemal
Length=4462
Score = 31.6 bits (70), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 20/89 (22%), Positives = 47/89 (52%), Gaps = 1/89 (1%)
Query 55 VEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQL 114
+E RR+ +VTP ++L + G+ + + + + + GL K+ + +V M +L
Sbjct 3004 LELRRYNYVTPTNYLELVSGYKKLLGEKRQELLDQANKLRTGLFKIDETREKVEVMSLEL 3063
Query 115 EKLQPELAKSSMETQQLMQVLTVKQEHAA 143
E + ++A+ + ++ + V+ V+Q+ A
Sbjct 3064 EDAKKKVAEFQKQCEEYL-VIIVQQKREA 3091
> hsa:1778 DYNC1H1, DHC1, DHC1a, DKFZp686P2245, DNCH1, DNCL, DNECL,
DYHC, Dnchc1, HL-3, KIAA0325, p22; dynein, cytoplasmic
1, heavy chain 1; K10413 dynein heavy chain 1, cytosolic
Length=4646
Score = 31.6 bits (70), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 20/83 (24%), Positives = 44/83 (53%), Gaps = 1/83 (1%)
Query 59 RFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQ 118
R +TP +L F++ + +F + S+ +Q +GL+K+ + QV ++++ L
Sbjct 3167 RTMAITPRHYLDFINHYANLFHEKRSELEEQQMHLNVGLRKIKETVDQVEELRRDLRIKS 3226
Query 119 PEL-AKSSMETQQLMQVLTVKQE 140
EL K++ +L +++ +QE
Sbjct 3227 QELEVKNAAANDKLKKMVKDQQE 3249
> mmu:13424 Dync1h1, 9930018I23Rik, AI894280, DHC1, DHC1a, DNCL,
Dnchc1, Dnec1, Dnecl, Loa, MAP1C, P22, Swl, mKIAA0325; dynein
cytoplasmic 1 heavy chain 1; K10413 dynein heavy chain
1, cytosolic
Length=4644
Score = 31.2 bits (69), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 20/83 (24%), Positives = 44/83 (53%), Gaps = 1/83 (1%)
Query 59 RFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQ 118
R +TP +L F++ + +F + S+ +Q +GL+K+ + QV ++++ L
Sbjct 3165 RTMAITPRHYLDFINHYANLFHEKRSELEEQQMHLNVGLRKIKETVDQVEELRRDLRIKS 3224
Query 119 PEL-AKSSMETQQLMQVLTVKQE 140
EL K++ +L +++ +QE
Sbjct 3225 QELEVKNAAANDKLKKMVKDQQE 3247
> cel:T21E12.4 dhc-1; Dynein Heavy Chain family member (dhc-1);
K10413 dynein heavy chain 1, cytosolic
Length=4568
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 0/63 (0%)
Query 59 RFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHDVSMQVMDMQQQLEKLQ 118
R TP FL F+ F +F + S ++ +GL K+ + QV ++Q+ L+
Sbjct 3110 RVMACTPRHFLDFIKQFMSLFHEKRSDLEEEKIHLNIGLNKISETEEQVKELQKSLKLKS 3169
Query 119 PEL 121
EL
Sbjct 3170 NEL 3172
> tgo:TGME49_061020 axonemal dynein heavy chain, putative (EC:2.4.1.14
1.1.1.6 1.1.1.47 1.6.5.3)
Length=4154
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 16/74 (21%), Positives = 34/74 (45%), Gaps = 0/74 (0%)
Query 43 FESTKNIADCYRVEQRRFFHVTPASFLRFLDGFCYVFMTRASQHHHKQQQYELGLQKLHD 102
F S A Y ++R+ ++TP +F+ + + + K+ + GL+KL D
Sbjct 2444 FYSVNEAAQEYLANEKRYAYITPKTFIESIKLYGSMLQENIDSLKSKRDRLSSGLRKLID 2503
Query 103 VSMQVMDMQQQLEK 116
+V ++ L++
Sbjct 2504 TREKVSALEDDLKE 2517
> sce:YKR054C DYN1, DHC1, PAC6; Cytoplasmic heavy chain dynein,
microtubule motor protein, required for anaphase spindle elongation;
involved in spindle assembly, chromosome movement,
and spindle orientation during cell division, targeted to
microtubule tips by Pac1p; K10413 dynein heavy chain 1, cytosolic
Length=4092
Score = 29.3 bits (64), Expect = 4.5, Method: Composition-based stats.
Identities = 21/89 (23%), Positives = 44/89 (49%), Gaps = 3/89 (3%)
Query 57 QRRFFHVTPASFLRFLDGFCYV--FMTRASQHHHKQQQY-ELGLQKLHDVSMQVMDMQQQ 113
Q+ V P S F+DG + +T Q + Q++ +GL+KL++ ++V ++ +
Sbjct 2978 QKMKVGVNPRSPGYFIDGLRALVKLVTAKYQDLQENQRFVNVGLEKLNESVLKVNELNKT 3037
Query 114 LEKLQPELAKSSMETQQLMQVLTVKQEHA 142
L K EL + E + + + ++Q +
Sbjct 3038 LSKKSTELTEKEKEARSTLDKMLMEQNES 3066
> ath:AT2G23140 binding / ubiquitin-protein ligase
Length=829
Score = 28.1 bits (61), Expect = 9.7, Method: Composition-based stats.
Identities = 30/122 (24%), Positives = 56/122 (45%), Gaps = 11/122 (9%)
Query 14 ADGTKESQLYPSADSNENLKHLCGACADIFESTKNIADCYRVEQRRFFHVTPASFLRFLD 73
A T ++ +P AD+NEN + A +++ I T A+ R L
Sbjct 447 ATSTVSNEEFPRADANENSEESAHATPYSSDASGEI------RSGPLAATTSAATRRDLS 500
Query 74 GFCYVFMTRASQHHHKQQQYE-LGLQKL----HDVSMQVMDMQQQLEKLQPELAKSSMET 128
F FM R ++ ++ E LG + + ++ + +++ Q++KL EL SS++T
Sbjct 501 DFSPKFMDRRTRGQFWRRPSERLGSRIVSAPSNETRRDLSEVETQVKKLVEELKSSSLDT 560
Query 129 QQ 130
Q+
Sbjct 561 QR 562
Lambda K H
0.320 0.129 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3068761412
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40