bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2307_orf1
Length=104
Score E
Sequences producing significant alignments: (Bits) Value
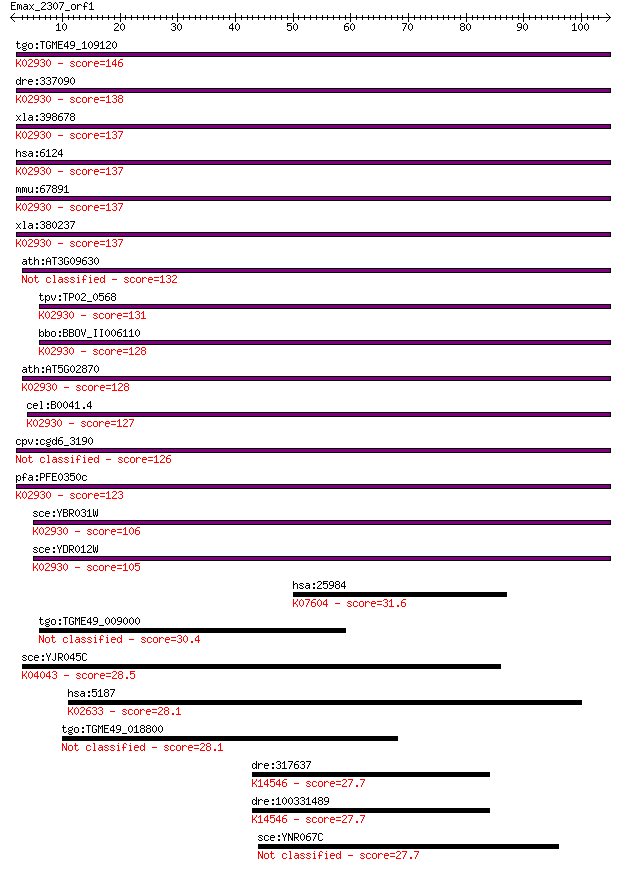
tgo:TGME49_109120 60s ribosomal protein L4, putative ; K02930 ... 146 1e-35
dre:337090 rpl4, fa97a02, fl04b10, wu:fa97a02, wu:fa97c01, wu:... 138 4e-33
xla:398678 rpl4-b, MGC64318, l1b, rpl1b; ribosomal protein L4;... 137 6e-33
hsa:6124 RPL4; ribosomal protein L4; K02930 large subunit ribo... 137 8e-33
mmu:67891 Rpl4, 2010004J23Rik; ribosomal protein L4; K02930 la... 137 1e-32
xla:380237 rpl4-a, MGC130828, MGC53834, rpl-4, rpl1a; ribosoma... 137 1e-32
ath:AT3G09630 60S ribosomal protein L4/L1 (RPL4A) 132 2e-31
tpv:TP02_0568 60S ribosomal protein L4/L1; K02930 large subuni... 131 5e-31
bbo:BBOV_II006110 18.m06507; ribosomal protein RPL4A; K02930 l... 128 5e-30
ath:AT5G02870 60S ribosomal protein L4/L1 (RPL4D); K02930 larg... 128 5e-30
cel:B0041.4 rpl-4; Ribosomal Protein, Large subunit family mem... 127 8e-30
cpv:cgd6_3190 60S ribosomal protein-like 126 1e-29
pfa:PFE0350c 60S ribosomal protein L4, putative; K02930 large ... 123 2e-28
sce:YBR031W RPL4A; Rpl4ap; K02930 large subunit ribosomal prot... 106 2e-23
sce:YDR012W RPL4B; Rpl4bp; K02930 large subunit ribosomal prot... 105 2e-23
hsa:25984 KRT23, CK23, DKFZp434G032, HAIK1, K23, MGC26158; ker... 31.6 0.57
tgo:TGME49_009000 HECT-domain (ubiquitin-transferase) containi... 30.4 1.6
sce:YJR045C SSC1, ENS1; Hsp70 family ATPase, constituent of th... 28.5 5.7
hsa:5187 PER1, MGC88021, PER, RIGUI, hPER; period homolog 1 (D... 28.1 6.2
tgo:TGME49_018800 hypothetical protein 28.1 6.4
dre:317637 wdr43, MGC110501, MGC172003, MGC174198, chunp6935, ... 27.7 9.1
dre:100331489 WD repeat domain 43-like; K14546 U3 small nucleo... 27.7 9.1
sce:YNR067C DSE4, ENG1; Daughter cell-specific secreted protei... 27.7 9.9
> tgo:TGME49_109120 60s ribosomal protein L4, putative ; K02930
large subunit ribosomal protein L4e
Length=416
Score = 146 bits (369), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 69/103 (66%), Positives = 82/103 (79%), Gaps = 2/103 (1%)
Query 2 MSATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
M+ RPLV VY PE S + L+P+V SPLRPDLVRFV+ N+ KN+RQ Y V+P A
Sbjct 1 MATARPLVSVYKPEDG--TASGTSLMPSVFLSPLRPDLVRFVHTNMAKNRRQPYGVAPNA 58
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
GY++SAESWGTGRAVSR+PRV GGGTHR+GQA+F NMCRGGGM
Sbjct 59 GYQTSAESWGTGRAVSRIPRVPGGGTHRAGQAAFGNMCRGGGM 101
> dre:337090 rpl4, fa97a02, fl04b10, wu:fa97a02, wu:fa97c01, wu:fl04b10,
zgc:56659, zgc:85693; ribosomal protein L4; K02930
large subunit ribosomal protein L4e
Length=375
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 62/103 (60%), Positives = 82/103 (79%), Gaps = 2/103 (1%)
Query 2 MSATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
M+ RPL+ VY+ +GE ++++PAV +P+RPD+V FV+ N+ KNKRQ YAVS A
Sbjct 1 MACARPLISVYSEKGE--TSGKNVVMPAVFRAPIRPDIVNFVHTNMRKNKRQPYAVSELA 58
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
G+++SAESWGTGRAV+R+PRV+GGGTHRSGQ +F NMCRGG M
Sbjct 59 GHQTSAESWGTGRAVARIPRVRGGGTHRSGQGAFGNMCRGGRM 101
> xla:398678 rpl4-b, MGC64318, l1b, rpl1b; ribosomal protein L4;
K02930 large subunit ribosomal protein L4e
Length=401
Score = 137 bits (346), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 61/103 (59%), Positives = 80/103 (77%), Gaps = 2/103 (1%)
Query 2 MSATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
M+ RPL+ VY+ +GE ++ +PAV +P+RPD+V FV+ N+ KN RQ YAVS A
Sbjct 5 MACARPLISVYSEKGE--TSGKNVTMPAVFKAPIRPDIVNFVHTNLRKNNRQPYAVSKLA 62
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
G+++SAESWGTGRAV+R+PRV+GGGTHRSGQ +F NMCRGG M
Sbjct 63 GHQTSAESWGTGRAVARIPRVRGGGTHRSGQGAFGNMCRGGRM 105
> hsa:6124 RPL4; ribosomal protein L4; K02930 large subunit ribosomal
protein L4e
Length=427
Score = 137 bits (345), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 62/103 (60%), Positives = 80/103 (77%), Gaps = 2/103 (1%)
Query 2 MSATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
M+ RPL+ VY+ +GE ++ LPAV +P+RPD+V FV+ N+ KN RQ YAVS A
Sbjct 1 MACARPLISVYSEKGE--SSGKNVTLPAVFKAPIRPDIVNFVHTNLRKNNRQPYAVSELA 58
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
G+++SAESWGTGRAV+R+PRV+GGGTHRSGQ +F NMCRGG M
Sbjct 59 GHQTSAESWGTGRAVARIPRVRGGGTHRSGQGAFGNMCRGGRM 101
> mmu:67891 Rpl4, 2010004J23Rik; ribosomal protein L4; K02930
large subunit ribosomal protein L4e
Length=419
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 62/103 (60%), Positives = 80/103 (77%), Gaps = 2/103 (1%)
Query 2 MSATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
M+ RPL+ VY+ +GE ++ LPAV +P+RPD+V FV+ N+ KN RQ YAVS A
Sbjct 1 MACARPLISVYSEKGE--SSGKNVTLPAVFKAPIRPDIVNFVHTNLRKNNRQPYAVSELA 58
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
G+++SAESWGTGRAV+R+PRV+GGGTHRSGQ +F NMCRGG M
Sbjct 59 GHQTSAESWGTGRAVARIPRVRGGGTHRSGQGAFGNMCRGGRM 101
> xla:380237 rpl4-a, MGC130828, MGC53834, rpl-4, rpl1a; ribosomal
protein L4; K02930 large subunit ribosomal protein L4e
Length=396
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 61/103 (59%), Positives = 80/103 (77%), Gaps = 2/103 (1%)
Query 2 MSATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
M+ RPL+ VY+ +GE ++ +PAV +P+RPD+V FV+ N+ KN RQ YAVS A
Sbjct 5 MACARPLISVYSEKGE--SSGKNVTMPAVFRAPIRPDIVNFVHTNLRKNNRQPYAVSKLA 62
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
G+++SAESWGTGRAV+R+PRV+GGGTHRSGQ +F NMCRGG M
Sbjct 63 GHQTSAESWGTGRAVARIPRVRGGGTHRSGQGAFGNMCRGGRM 105
> ath:AT3G09630 60S ribosomal protein L4/L1 (RPL4A)
Length=405
Score = 132 bits (332), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 62/103 (60%), Positives = 81/103 (78%), Gaps = 1/103 (0%)
Query 3 SATRPLVGVYTPEGER-LPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
+A RPLV + T +G+ SS+++LP V+++P+RPD+V FV+ I N RQ YAVS A
Sbjct 4 AAARPLVTIQTLDGDMSTDQSSTVVLPDVMTAPVRPDIVNFVHAQISNNSRQPYAVSKKA 63
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
G+++SAESWGTGRAVSR+PRV GGGTHR+GQA+F NMCRGG M
Sbjct 64 GHQTSAESWGTGRAVSRIPRVPGGGTHRAGQAAFGNMCRGGRM 106
> tpv:TP02_0568 60S ribosomal protein L4/L1; K02930 large subunit
ribosomal protein L4e
Length=389
Score = 131 bits (330), Expect = 5e-31, Method: Compositional matrix adjust.
Identities = 62/99 (62%), Positives = 78/99 (78%), Gaps = 2/99 (2%)
Query 6 RPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYAGYKS 65
RP+V V++ ER S ++P V S+PLR DLVR+V+ N+ KNKRQ+YAVS +GY++
Sbjct 4 RPVVSVFSVSTER--RMGSTVMPRVFSTPLRSDLVRYVHTNMSKNKRQAYAVSRLSGYQT 61
Query 66 SAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
SA SWGTGRA+SR PRVKGGGTHRSGQA++AN CR GGM
Sbjct 62 SARSWGTGRALSRTPRVKGGGTHRSGQAAYANFCRAGGM 100
> bbo:BBOV_II006110 18.m06507; ribosomal protein RPL4A; K02930
large subunit ribosomal protein L4e
Length=360
Score = 128 bits (321), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 61/100 (61%), Positives = 79/100 (79%), Gaps = 4/100 (4%)
Query 6 RPLVGVYTP-EGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYAGYK 64
RP V V+ +GER+ + L+P V S+PLR DLVR V+ N+ KN+RQ+YAVS +GY+
Sbjct 4 RPEVSVHNATDGERV---GATLMPKVFSAPLRLDLVRIVHTNMSKNRRQAYAVSKMSGYQ 60
Query 65 SSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
+SA SWGTGRA++RVPRVKGGGTHR+GQA++AN CR GGM
Sbjct 61 TSARSWGTGRAMARVPRVKGGGTHRAGQAAYANFCRAGGM 100
> ath:AT5G02870 60S ribosomal protein L4/L1 (RPL4D); K02930 large
subunit ribosomal protein L4e
Length=406
Score = 128 bits (321), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 61/103 (59%), Positives = 79/103 (76%), Gaps = 1/103 (0%)
Query 3 SATRPLVGVYTPEGER-LPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
+A RPLV V +G+ S+++ LP V+++P+RPD+V FV+ I N RQ YAVS A
Sbjct 5 AAARPLVTVQGLDGDMSTDQSTTVTLPDVMTAPVRPDIVNFVHAQISNNSRQPYAVSKKA 64
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
G+++SAESWGTGRAVSR+PRV GGGTHR+GQA+F NMCRGG M
Sbjct 65 GHQTSAESWGTGRAVSRIPRVPGGGTHRAGQAAFGNMCRGGRM 107
> cel:B0041.4 rpl-4; Ribosomal Protein, Large subunit family member
(rpl-4); K02930 large subunit ribosomal protein L4e
Length=345
Score = 127 bits (319), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 60/101 (59%), Positives = 76/101 (75%), Gaps = 2/101 (1%)
Query 4 ATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYAGY 63
A RPLV VY + E S I LPAV +P+RPDLV F+ + + +N+RQ++AV+ AG
Sbjct 2 AARPLVTVYDEKYE--ATQSQIRLPAVFRTPIRPDLVSFIADQVRRNRRQAHAVNTKAGK 59
Query 64 KSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
+ SAESWGTGRAV+R+PRV+GGGTHRSGQ +F NMCRGG M
Sbjct 60 QHSAESWGTGRAVARIPRVRGGGTHRSGQGAFGNMCRGGHM 100
> cpv:cgd6_3190 60S ribosomal protein-like
Length=394
Score = 126 bits (317), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 61/103 (59%), Positives = 75/103 (72%), Gaps = 1/103 (0%)
Query 2 MSATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
M+ RP+V V+ P + +SSI LP V SPLR DLVR+V+ N+ KNKRQ+Y VS A
Sbjct 1 MTFARPIVSVF-PSNDSTKATSSIPLPTVFVSPLRHDLVRYVFTNMSKNKRQAYGVSTKA 59
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
G++ SAESWGTGRAV+R+PRV GT RSGQ +F NMCRGG M
Sbjct 60 GHQHSAESWGTGRAVARIPRVSASGTSRSGQGAFGNMCRGGHM 102
> pfa:PFE0350c 60S ribosomal protein L4, putative; K02930 large
subunit ribosomal protein L4e
Length=411
Score = 123 bits (308), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/103 (54%), Positives = 75/103 (72%), Gaps = 2/103 (1%)
Query 2 MSATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYA 61
M+ RP+ VY+ G+ + + +P V +P+R DL++ VY N+ KN+R YAV A
Sbjct 1 MATIRPVANVYSTNGKNV--VGEVEIPVVFQTPIRNDLIQSVYTNMSKNRRHPYAVKLGA 58
Query 62 GYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
GY++SAESWGTGRAV+R+PRV GGGTHR+GQ +F NMCRGGGM
Sbjct 59 GYETSAESWGTGRAVARIPRVPGGGTHRAGQGAFGNMCRGGGM 101
> sce:YBR031W RPL4A; Rpl4ap; K02930 large subunit ribosomal protein
L4e
Length=362
Score = 106 bits (264), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 59/100 (59%), Positives = 79/100 (79%), Gaps = 2/100 (2%)
Query 5 TRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYAGYK 64
+RP V V++ GE ++++ LPAV S+P+RPD+V V+ ++ KNKRQ+YAVS AG++
Sbjct 2 SRPQVTVHSLTGE--ATANALPLPAVFSAPIRPDIVHTVFTSVNKNKRQAYAVSEKAGHQ 59
Query 65 SSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
+SAESWGTGRAV+R+PRV GGGT RSGQ +F NMCRGG M
Sbjct 60 TSAESWGTGRAVARIPRVGGGGTGRSGQGAFGNMCRGGRM 99
> sce:YDR012W RPL4B; Rpl4bp; K02930 large subunit ribosomal protein
L4e
Length=362
Score = 105 bits (263), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 59/100 (59%), Positives = 79/100 (79%), Gaps = 2/100 (2%)
Query 5 TRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYAGYK 64
+RP V V++ GE ++++ LPAV S+P+RPD+V V+ ++ KNKRQ+YAVS AG++
Sbjct 2 SRPQVTVHSLTGE--ATANALPLPAVFSAPIRPDIVHTVFTSVNKNKRQAYAVSEKAGHQ 59
Query 65 SSAESWGTGRAVSRVPRVKGGGTHRSGQASFANMCRGGGM 104
+SAESWGTGRAV+R+PRV GGGT RSGQ +F NMCRGG M
Sbjct 60 TSAESWGTGRAVARIPRVGGGGTGRSGQGAFGNMCRGGRM 99
> hsa:25984 KRT23, CK23, DKFZp434G032, HAIK1, K23, MGC26158; keratin
23 (histone deacetylase inducible); K07604 type I keratin,
acidic
Length=422
Score = 31.6 bits (70), Expect = 0.57, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 18/37 (48%), Gaps = 0/37 (0%)
Query 50 NKRQSYAVSPYAGYKSSAESWGTGRAVSRVPRVKGGG 86
N S++ +P A + + WG R+ R P V GG
Sbjct 2 NSGHSFSQTPSASFHGAGGGWGRPRSFPRAPTVHGGA 38
> tgo:TGME49_009000 HECT-domain (ubiquitin-transferase) containing
protein (EC:3.1.3.18 2.7.7.19 3.4.21.10 2.1.3.1)
Length=10999
Score = 30.4 bits (67), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/53 (33%), Positives = 27/53 (50%), Gaps = 1/53 (1%)
Query 6 RPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVS 58
R V PEG+R+PP LP + R L R+ N+I + +R YA++
Sbjct 10926 RMAVDYEIPEGQRVPPIDDNRLPTAATCGFRLLLPRYTSNSIFR-QRLLYAIN 10977
> sce:YJR045C SSC1, ENS1; Hsp70 family ATPase, constituent of
the import motor component of the Translocase of the Inner Mitochondrial
membrane (TIM23 complex); involved in protein translocation
and folding; subunit of SceI endonuclease; K04043
molecular chaperone DnaK
Length=654
Score = 28.5 bits (62), Expect = 5.7, Method: Composition-based stats.
Identities = 25/90 (27%), Positives = 38/90 (42%), Gaps = 12/90 (13%)
Query 3 SATRPLVGVYTPEGERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVS---- 58
S T P V +T EGERL + +PA + + P+ F +I + + V
Sbjct 61 SRTTPSVVAFTKEGERL-----VGIPAKRQAVVNPENTLFATKRLIGRRFEDAEVQRDIK 115
Query 59 --PYAGYK-SSAESWGTGRAVSRVPRVKGG 85
PY K S+ ++W R + P GG
Sbjct 116 QVPYKIVKHSNGDAWVEARGQTYSPAQIGG 145
> hsa:5187 PER1, MGC88021, PER, RIGUI, hPER; period homolog 1
(Drosophila); K02633 period circadian protein
Length=1290
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 41/94 (43%), Gaps = 8/94 (8%)
Query 11 VYTPEG--ERLPPSSSILLPAVLSSPLRPDLVRFVYNNIIKNKRQSYAVSPYAGYKSSAE 68
V++P G + LPP+ + + PA +PL +V V N + SY PY ++ AE
Sbjct 888 VFSPRGGPQPLPPAPTSVPPAAFPAPLVTPMVALVLPNYLFPTPSSY---PYGALQTPAE 944
Query 69 SWGTGRAVS---RVPRVKGGGTHRSGQASFANMC 99
T + S +P + HR F + C
Sbjct 945 GPPTPASHSPSPSLPALAPSPPHRPDSPLFNSRC 978
> tgo:TGME49_018800 hypothetical protein
Length=1196
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 36/79 (45%), Gaps = 24/79 (30%)
Query 10 GVYTPE---------GERLPPSSSI------------LLPAVLSSPLRPDLVRFVYNNII 48
GV TPE R PPS+S L PAV + RP+ V N +
Sbjct 94 GVCTPEPSDSAAQGSRAREPPSASAGDSQATCRASGRLQPAVAARERRPESVEPRGRNFV 153
Query 49 KNKRQSYAVSPYAGYKSSA 67
K++RQ +SP AG K+S+
Sbjct 154 KSERQ---LSPSAGNKASS 169
> dre:317637 wdr43, MGC110501, MGC172003, MGC174198, chunp6935,
kiaa0007l, wdr43l, wu:fb83h12, wu:fc13e01, zgc:110501; WD
repeat domain 43; K14546 U3 small nucleolar RNA-associated protein
5
Length=650
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 43 VYNNIIKNKRQSYAVSPYAGYKSSAESWGTGRAVSRVPRVK 83
++N K +Q Y S + + +WG RAV VP+ K
Sbjct 33 IWNTESKTLQQEYVPSAHLSAACTCVTWGPCRAVQEVPQRK 73
> dre:100331489 WD repeat domain 43-like; K14546 U3 small nucleolar
RNA-associated protein 5
Length=650
Score = 27.7 bits (60), Expect = 9.1, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 43 VYNNIIKNKRQSYAVSPYAGYKSSAESWGTGRAVSRVPRVK 83
++N K +Q Y S + + +WG RAV VP+ K
Sbjct 33 IWNTESKTLQQEYVPSAHLSAACTCVTWGPCRAVQEVPQRK 73
> sce:YNR067C DSE4, ENG1; Daughter cell-specific secreted protein
with similarity to glucanases, degrades cell wall from the
daughter side causing daughter to separate from mother (EC:3.2.1.6)
Length=1117
Score = 27.7 bits (60), Expect = 9.9, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 44 YNNIIKNKRQSYA-VSPYAGYKSSAESWGTGRAVSRVPRVKGGGTHRSGQASF 95
Y N+I ++S A V PY+ +K ++ S+G + V + GG SG A +
Sbjct 440 YTNLIVGSQESPAFVYPYSLWKYTSSSYGFAVQHTTVDQYSYGGYDSSGNAEY 492
Lambda K H
0.315 0.132 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2022291452
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40