bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2325_orf1
Length=130
Score E
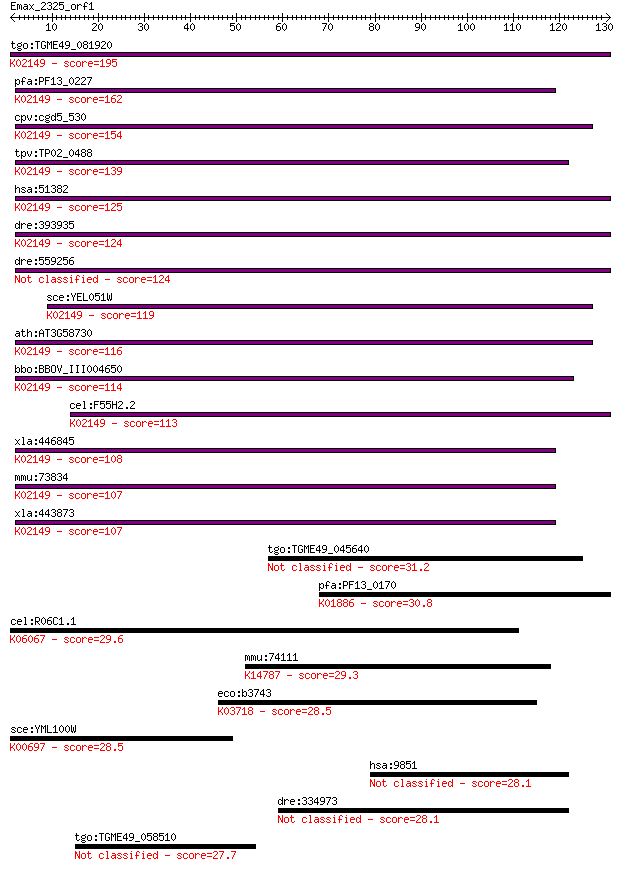
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_081920 vacuolar ATP synthase subunit D, putative (E... 195 3e-50
pfa:PF13_0227 vacuolar ATP synthase subunit D, putative (EC:3.... 162 2e-40
cpv:cgd5_530 vacuolar H-ATpase subunit D ; K02149 V-type H+-tr... 154 5e-38
tpv:TP02_0488 vacuolar ATP synthase subunit D; K02149 V-type H... 139 2e-33
hsa:51382 ATP6V1D, ATP6M, VATD, VMA8; ATPase, H+ transporting,... 125 4e-29
dre:393935 atp6v1d, MGC55524, zgc:55524; ATPase, H+ transporti... 124 6e-29
dre:559256 atp6v1d; ATPase, H+ transporting, V1 subunit D 124 6e-29
sce:YEL051W VMA8; Subunit D of the eight-subunit V1 peripheral... 119 3e-27
ath:AT3G58730 vacuolar ATP synthase subunit D (VATD) / V-ATPas... 116 2e-26
bbo:BBOV_III004650 17.m07416; V-type ATPase, D subunit family ... 114 1e-25
cel:F55H2.2 vha-14; Vacuolar H ATPase family member (vha-14); ... 113 1e-25
xla:446845 atp6v1d, MGC80692; ATPase, H+ transporting, lysosom... 108 5e-24
mmu:73834 Atp6v1d, 1110004P10Rik, Atp6m, VATD, Vma8; ATPase, H... 107 7e-24
xla:443873 MGC79146 protein; K02149 V-type H+-transporting ATP... 107 1e-23
tgo:TGME49_045640 hypothetical protein 31.2 0.89
pfa:PF13_0170 glutaminyl-tRNA synthetase, putative (EC:6.1.1.1... 30.8 1.1
cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3); ... 29.6 2.4
mmu:74111 Rbm19, 1200009A02Rik, AV302714, KIAA0682, NPO; RNA b... 29.3 3.0
eco:b3743 asnC, ECK3737, JW3721; DNA-binding transcriptional d... 28.5 5.9
sce:YML100W TSL1; Large subunit of trehalose 6-phosphate synth... 28.5 6.1
hsa:9851 MGC130040, MGC130041; KIAA0753 28.1 7.4
dre:334973 ttf1, wu:fa11a12; transcription termination factor,... 28.1 7.7
tgo:TGME49_058510 cAMP-specific phosphodiesterase, putative (E... 27.7 9.8
> tgo:TGME49_081920 vacuolar ATP synthase subunit D, putative
(EC:3.6.3.14); K02149 V-type H+-transporting ATPase subunit
D [EC:3.6.3.14]
Length=245
Score = 195 bits (496), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 90/130 (69%), Positives = 111/130 (85%), Gaps = 0/130 (0%)
Query 1 RIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAE 60
R++RPA F+ A DNVAGV LP+F I TDP+VD+LKN+ ++AGG VI+AAR+ + + +E
Sbjct 84 RVRRPATFLNVAADNVAGVTLPIFHICTDPSVDVLKNVGVAAGGQVIMAAREMFLKVFSE 143
Query 61 LVKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKK 120
LVKLASLQTAFFTLD EIKMTNRRVNAL+NVVLP++D I YI +ELDEMEREEFFRLKK
Sbjct 144 LVKLASLQTAFFTLDEEIKMTNRRVNALSNVVLPRIDGGINYIVRELDEMEREEFFRLKK 203
Query 121 IQEKKKVARE 130
IQEKK++ +E
Sbjct 204 IQEKKRMRKE 213
> pfa:PF13_0227 vacuolar ATP synthase subunit D, putative (EC:3.6.3.14);
K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 162 bits (411), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 75/117 (64%), Positives = 94/117 (80%), Gaps = 0/117 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
IKRP V ++ + +NVAGV+LP+FQ+ DPTVD+L N+ ++AGG VI R+ Y + L L
Sbjct 87 IKRPVVTLSLSTNNVAGVKLPIFQVNIDPTVDVLGNLGVAAGGQVINNTRENYLQCLNML 146
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRL 118
VKLAS+Q AFF+LD EIKMTNRRVNALNN+VLP+LD I YI KELDE+EREEF+RL
Sbjct 147 VKLASMQVAFFSLDEEIKMTNRRVNALNNIVLPRLDGGINYIIKELDEIEREEFYRL 203
> cpv:cgd5_530 vacuolar H-ATpase subunit D ; K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=249
Score = 154 bits (390), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 76/126 (60%), Positives = 95/126 (75%), Gaps = 1/126 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDI-LKNINLSAGGHVILAARDKYQEALAE 60
KRP V + +N+AGVRLP+F++ D +I +++GG VI + R+ Y + L +
Sbjct 78 CKRPTVTMEVGTENIAGVRLPIFEMNVDNNSSTETCHIGVASGGQVIQSTREIYMKVLRD 137
Query 61 LVKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKK 120
LVKLASLQTAFF+LD EIKMTNRRVNAL NVVLPKL+ + YI +ELDE+EREEFFRLKK
Sbjct 138 LVKLASLQTAFFSLDEEIKMTNRRVNALQNVVLPKLEDGMNYILRELDEIEREEFFRLKK 197
Query 121 IQEKKK 126
IQEKKK
Sbjct 198 IQEKKK 203
> tpv:TP02_0488 vacuolar ATP synthase subunit D; K02149 V-type
H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=238
Score = 139 bits (351), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 65/120 (54%), Positives = 91/120 (75%), Gaps = 0/120 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ RP+V + +N+AGV LPVF + TDPTVD+ N++LS+GG I + + + AL L
Sbjct 88 VGRPSVTLKLRGENIAGVLLPVFSLQTDPTVDLFANLSLSSGGSAIQSVKTTHLAALDIL 147
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
V+LASLQ +F L+ EI+MTNRR+NAL+NV++P +DR++ YI +ELDEMEREEF+RLK I
Sbjct 148 VELASLQISFIILNEEIRMTNRRINALDNVLIPSIDRNLEYIRRELDEMEREEFYRLKMI 207
> hsa:51382 ATP6V1D, ATP6M, VATD, VMA8; ATPase, H+ transporting,
lysosomal 34kDa, V1 subunit D (EC:3.6.3.14 3.6.1.34); K02149
V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 125 bits (313), Expect = 4e-29, Method: Compositional matrix adjust.
Identities = 65/129 (50%), Positives = 91/129 (70%), Gaps = 1/129 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ + V + A DNVAGV LPVF+ + T D + L+ GG + + Y +A+ L
Sbjct 87 VNKAQVKIRAKKDNVAGVTLPVFEHYHEGT-DSYELTGLARGGEQLAKLKRNYAKAVELL 145
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
V+LASLQT+F TLD IK+TNRRVNA+ +V++P+++R++ YI ELDE EREEF+RLKKI
Sbjct 146 VELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLAYIITELDEREREEFYRLKKI 205
Query 122 QEKKKVARE 130
QEKKK+ +E
Sbjct 206 QEKKKILKE 214
> dre:393935 atp6v1d, MGC55524, zgc:55524; ATPase, H+ transporting,
V1 subunit D; K02149 V-type H+-transporting ATPase subunit
D [EC:3.6.3.14]
Length=248
Score = 124 bits (312), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 67/129 (51%), Positives = 90/129 (69%), Gaps = 1/129 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ + V V A DNVAGV LPVF+ + D + L+ GG + + Y +A+ L
Sbjct 87 VNKAQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLARGGEQLSRLKRNYAKAVELL 145
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
V+LASLQT+F TLD IK+TNRRVNA+ +V++P+++R++TYI ELDE EREEF+RLKKI
Sbjct 146 VELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLTYIITELDEREREEFYRLKKI 205
Query 122 QEKKKVARE 130
QEKKK RE
Sbjct 206 QEKKKQLRE 214
> dre:559256 atp6v1d; ATPase, H+ transporting, V1 subunit D
Length=248
Score = 124 bits (312), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 67/129 (51%), Positives = 90/129 (69%), Gaps = 1/129 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ + V V A DNVAGV LPVF+ + D + L+ GG + + Y +A+ L
Sbjct 87 VNKAQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLARGGEQLSRLKRNYAKAVELL 145
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
V+LASLQT+F TLD IK+TNRRVNA+ +V++P+++R++TYI ELDE EREEF+RLKKI
Sbjct 146 VELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLTYIITELDEREREEFYRLKKI 205
Query 122 QEKKKVARE 130
QEKKK RE
Sbjct 206 QEKKKQLRE 214
> sce:YEL051W VMA8; Subunit D of the eight-subunit V1 peripheral
membrane domain of the vacuolar H+-ATPase (V-ATPase), an
electrogenic proton pump found throughout the endomembrane system;
plays a role in the coupling of proton transport and
ATP hydrolysis (EC:3.6.3.14); K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=256
Score = 119 bits (297), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 58/118 (49%), Positives = 82/118 (69%), Gaps = 0/118 (0%)
Query 9 VTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQ 68
V A +NV+GV L F+ DP ++ + L GG + A++ Y A+ LV+LASLQ
Sbjct 94 VRARQENVSGVYLSQFESYIDPEINDFRLTGLGRGGQQVQRAKEIYSRAVETLVELASLQ 153
Query 69 TAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKIQEKKK 126
TAF LD IK+TNRRVNA+ +V++P+ + +I YI ELDE++REEF+RLKK+QEKK+
Sbjct 154 TAFIILDEVIKVTNRRVNAIEHVIIPRTENTIAYINSELDELDREEFYRLKKVQEKKQ 211
> ath:AT3G58730 vacuolar ATP synthase subunit D (VATD) / V-ATPase
D subunit / vacuolar proton pump D subunit (VATPD); K02149
V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=261
Score = 116 bits (291), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 65/126 (51%), Positives = 85/126 (67%), Gaps = 3/126 (2%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVF-QITTDPTVDILKNINLSAGGHVILAARDKYQEALAE 60
+K + V + +N+AGV+LP F + T + L L+ GG + A R Y +A+
Sbjct 89 VKEATLKVRSRTENIAGVKLPKFDHFSEGETKNDL--TGLARGGQQVRACRVAYVKAIEV 146
Query 61 LVKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKK 120
LV+LASLQT+F TLD IK TNRRVNAL NVV PKL+ +I+YI ELDE+ERE+FFRLKK
Sbjct 147 LVELASLQTSFLTLDEAIKTTNRRVNALENVVKPKLENTISYIKGELDELEREDFFRLKK 206
Query 121 IQEKKK 126
IQ K+
Sbjct 207 IQGYKR 212
> bbo:BBOV_III004650 17.m07416; V-type ATPase, D subunit family
protein; K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=233
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 63/121 (52%), Positives = 89/121 (73%), Gaps = 0/121 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ R AV + +NVAGV +P F++ DPTVD++ NI L+ GGHVI + + + E L L
Sbjct 73 VGRSAVTLRVRTENVAGVIIPHFELKIDPTVDVIANIGLTTGGHVIHSVKTAHLEFLETL 132
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
+LASLQ +F L+ EIKMTNRRVNAL+N+V+P +D ++ YI +ELDE+EREEF+RLK +
Sbjct 133 AELASLQVSFMMLEQEIKMTNRRVNALDNLVIPTIDNNLEYIKRELDELEREEFYRLKMV 192
Query 122 Q 122
+
Sbjct 193 R 193
> cel:F55H2.2 vha-14; Vacuolar H ATPase family member (vha-14);
K02149 V-type H+-transporting ATPase subunit D [EC:3.6.3.14]
Length=257
Score = 113 bits (283), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 60/117 (51%), Positives = 78/117 (66%), Gaps = 1/117 (0%)
Query 14 DNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQTAFFT 73
+NV GV LPVF D D L GG I + Y +A+ LV+LA+LQT F T
Sbjct 101 ENVVGVFLPVFDAYQDGP-DAYDLTGLGKGGANIARLKKNYNKAIELLVELATLQTCFIT 159
Query 74 LDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKIQEKKKVARE 130
LD IK+TNRRVNA+ +V++P+++ ++TYI ELDEMEREEFFR+KKIQ KK +E
Sbjct 160 LDEAIKVTNRRVNAIEHVIIPRIENTLTYIVTELDEMEREEFFRMKKIQANKKKLKE 216
> xla:446845 atp6v1d, MGC80692; ATPase, H+ transporting, lysosomal
34kDa, V1 subunit D (EC:3.6.3.14); K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=248
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 57/117 (48%), Positives = 80/117 (68%), Gaps = 1/117 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ + V V A DNVAGV LPVF+ + D + L+ GG + + Y +A+ L
Sbjct 87 VNKSQVKVRAKKDNVAGVTLPVFEHYQEGG-DSYELTGLARGGEQLAKLKRNYAKAVELL 145
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRL 118
V+LASLQT+F TLD IK+TNRRVNA+ +V++PK++R+++YI ELDE EREEF+RL
Sbjct 146 VELASLQTSFVTLDEAIKITNRRVNAIEHVIIPKIERTLSYIITELDEREREEFYRL 202
> mmu:73834 Atp6v1d, 1110004P10Rik, Atp6m, VATD, Vma8; ATPase,
H+ transporting, lysosomal V1 subunit D; K02149 V-type H+-transporting
ATPase subunit D [EC:3.6.3.14]
Length=247
Score = 107 bits (268), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 56/117 (47%), Positives = 80/117 (68%), Gaps = 1/117 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ + V + A DNVAGV LPVF+ + T D + L+ GG + + Y +A+ L
Sbjct 87 VNKAQVKIRAKKDNVAGVTLPVFEHYHEGT-DSYELTGLARGGEQLAKLKRNYAKAVELL 145
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRL 118
V+LASLQT+F TLD IK+TNRRVNA+ +V++P+++R++ YI ELDE EREEF+RL
Sbjct 146 VELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLAYIITELDEREREEFYRL 202
> xla:443873 MGC79146 protein; K02149 V-type H+-transporting ATPase
subunit D [EC:3.6.3.14]
Length=246
Score = 107 bits (266), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 56/117 (47%), Positives = 80/117 (68%), Gaps = 1/117 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ + V V A DNVAGV LPVF+ + D + L+ GG + + Y +A+ L
Sbjct 87 VNKAQVKVRARKDNVAGVTLPVFEHYQEGG-DSYELTGLARGGEQLAKLKRNYAKAVELL 145
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRL 118
V+LASLQT+F TLD IK+TNRRVNA+ +V++P+++R+++YI ELDE EREEF+RL
Sbjct 146 VELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLSYIVTELDEREREEFYRL 202
> tgo:TGME49_045640 hypothetical protein
Length=416
Score = 31.2 bits (69), Expect = 0.89, Method: Composition-based stats.
Identities = 19/68 (27%), Positives = 37/68 (54%), Gaps = 7/68 (10%)
Query 57 ALAELVKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFF 116
LAE ++ SL +L+ + ++ R+++A+ ++PKLDR+ T + + E+F
Sbjct 47 GLAESKRVPSLTQP--SLERSVTISRRKIDAVGMSLVPKLDRTTTSLAA-----KEEKFS 99
Query 117 RLKKIQEK 124
+ KI K
Sbjct 100 SIDKIVSK 107
> pfa:PF13_0170 glutaminyl-tRNA synthetase, putative (EC:6.1.1.18);
K01886 glutaminyl-tRNA synthetase [EC:6.1.1.18]
Length=918
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 38/67 (56%), Gaps = 6/67 (8%)
Query 68 QTAFFTLDA----EIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKIQE 123
+ FFT D E+ + N V ++N +L K + + KELD+++RE+ +K+++
Sbjct 847 RVGFFTKDKDTTNELPVFNLTVPLVDNTMLKKKKEDL--LQKELDKLKREKIAAERKLKK 904
Query 124 KKKVARE 130
++K RE
Sbjct 905 EQKKIRE 911
> cel:R06C1.1 hda-3; Histone DeAcetylase family member (hda-3);
K06067 histone deacetylase 1/2 [EC:3.5.1.98]
Length=465
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 32/124 (25%), Positives = 58/124 (46%), Gaps = 14/124 (11%)
Query 1 RIKRPAVFVTAAYDNVAGVRLPVFQITT---DPTVDILKNIN-----LSAGGHVILAARD 52
R + AV + D++AG RL VF +TT V+ +K+ N + GG+ I
Sbjct 250 RFQPEAVVLQCGADSLAGDRLGVFNLTTYGHGKCVEYMKSFNVPLLLVGGGGYTIRNVSR 309
Query 53 --KYQEALA---ELVKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPK-LDRSITYITKE 106
Y+ A+A E+ L F + K+ + + AL+N P+ +D++I + +
Sbjct 310 CWLYETAIALNQEVSDDLPLHDYFDYFIPDYKLHIKPLAALSNFNTPEFIDQTIVALLEN 369
Query 107 LDEM 110
L ++
Sbjct 370 LKQL 373
> mmu:74111 Rbm19, 1200009A02Rik, AV302714, KIAA0682, NPO; RNA
binding motif protein 19; K14787 multiple RNA-binding domain-containing
protein 1
Length=952
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 34/68 (50%), Gaps = 4/68 (5%)
Query 52 DKYQEALAELVKLASLQTAF--FTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDE 109
D + +A AE K L TL AEI+ T R +L V+LP + IT I + L+
Sbjct 572 DSFSQAAAERSKTVILAKNLPAGTLAAEIQETFSRFGSLGRVLLP--EGGITAIVEFLEP 629
Query 110 MEREEFFR 117
+E + FR
Sbjct 630 LEARKAFR 637
> eco:b3743 asnC, ECK3737, JW3721; DNA-binding transcriptional
dual regulator; K03718 Lrp/AsnC family transcriptional regulator,
regulator for asnA, asnC and gidA
Length=152
Score = 28.5 bits (62), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 37/72 (51%), Gaps = 11/72 (15%)
Query 46 VILAARDKYQEALAELVKLASLQTAFFTLDAE---IKMTNRRVNALNNVVLPKLDRSITY 102
+IL + Y ALA+L L + A++T IK+ R ++AL +V++ K+
Sbjct 74 IILKSAKDYPSALAKLESLDEVTEAYYTTGHYSIFIKVMCRSIDALQHVLINKI------ 127
Query 103 ITKELDEMEREE 114
+ +DE++ E
Sbjct 128 --QTIDEIQSTE 137
> sce:YML100W TSL1; Large subunit of trehalose 6-phosphate synthase
(Tps1p)/phosphatase (Tps2p) complex, which converts uridine-5'-diphosphoglucose
and glucose 6-phosphate to trehalose,
homologous to Tps3p and may share function; K00697 alpha,alpha-trehalose-phosphate
synthase (UDP-forming) [EC:2.4.1.15]
Length=1098
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 3/48 (6%)
Query 1 RIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVIL 48
RI P + +D+ + + P+ Q DPT ++LKN+N S H +L
Sbjct 144 RIASP---IQHEHDSGSRIASPIQQQQQDPTTNLLKNVNKSLLVHSLL 188
> hsa:9851 MGC130040, MGC130041; KIAA0753
Length=967
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 12/43 (27%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 79 KMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
++ + ++++ L LD + TKEL+E++ EE +RL+++
Sbjct 627 ELKAKEIDSMQKQRLDWLDAETSRRTKELNELKAEEMYRLQQL 669
> dre:334973 ttf1, wu:fa11a12; transcription termination factor,
RNA polymerase I
Length=577
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 30/63 (47%), Gaps = 0/63 (0%)
Query 59 AELVKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRL 118
A + K+ ++ D + V+ L+N +LP L RS ++E++E FRL
Sbjct 469 AHVQKMWHKMKNYYVPDWNSRSFKDNVSHLHNDILPCLVRSCKGFDVNAVQVEQKELFRL 528
Query 119 KKI 121
KI
Sbjct 529 SKI 531
> tgo:TGME49_058510 cAMP-specific phosphodiesterase, putative
(EC:3.1.4.17)
Length=951
Score = 27.7 bits (60), Expect = 9.8, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 15 NVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDK 53
N G +LP++Q+ +DIL N ++S H + R K
Sbjct 791 NFFGDKLPLWQVFRLHMIDILLNFDISKSAHQVAMFRTK 829
Lambda K H
0.320 0.135 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2039374804
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40