bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2408_orf1
Length=127
Score E
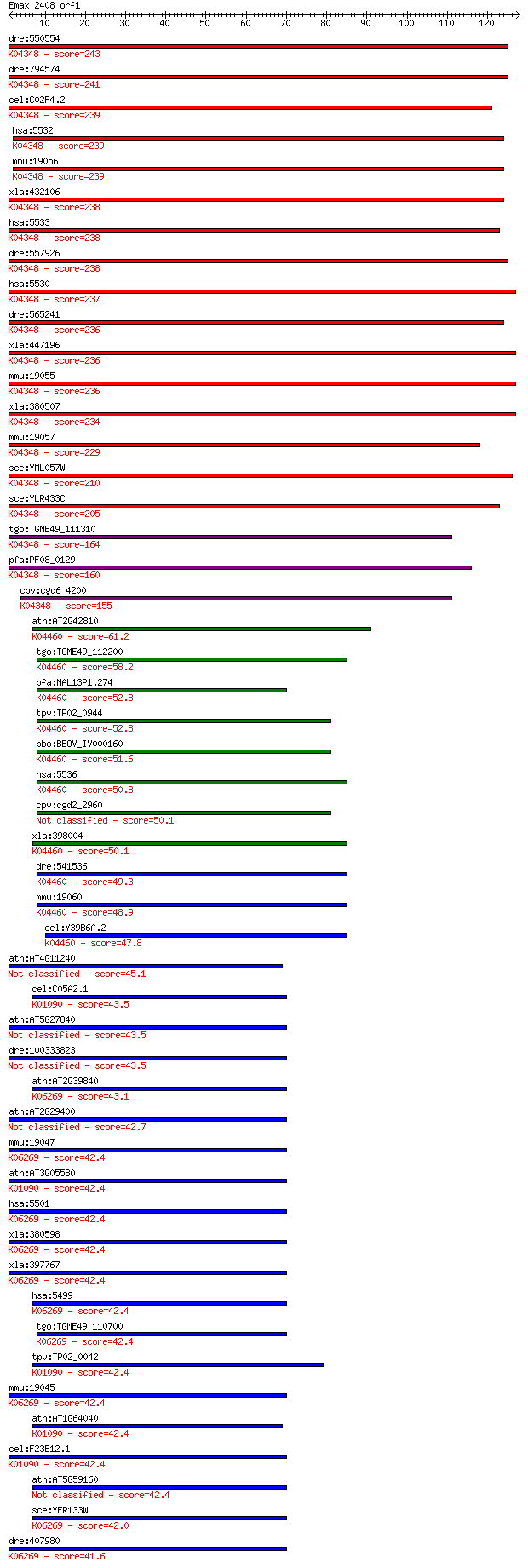
Sequences producing significant alignments: (Bits) Value
dre:550554 ppp3cb, wu:fq19c08, zgc:110141; protein phosphatase... 243 2e-64
dre:794574 ppp3ca, fj46e08, si:dkeyp-79c2.2, wu:fj46e08; prote... 241 3e-64
cel:C02F4.2 tax-6; abnormal CHEmotaxis family member (tax-6); ... 239 1e-63
hsa:5532 PPP3CB, CALNA2, CALNB, CNA2, PP2Bbeta; protein phosph... 239 2e-63
mmu:19056 Ppp3cb, 1110063J16Rik, Calnb, CnAbeta, Cnab; protein... 239 2e-63
xla:432106 ppp3cb, MGC81043, calna2, calnb, cna2, pp2bbeta; pr... 238 2e-63
hsa:5533 PPP3CC, CALNA3, CNA3, PP2Bgamma; protein phosphatase ... 238 3e-63
dre:557926 ppp3cca, ppp3cc, sb:cb767, si:dkey-21k4.5; protein ... 238 5e-63
hsa:5530 PPP3CA, CALN, CALNA, CALNA1, CCN1, CNA1, PPP2B; prote... 237 8e-63
dre:565241 ppp3ccb, im:7163784, zgc:158685; protein phosphatas... 236 1e-62
xla:447196 ppp3ca, MGC79144, calcineurin, caln, calna, calna1,... 236 1e-62
mmu:19055 Ppp3ca, 2900074D19Rik, AI841391, AW413465, CN, Caln,... 236 2e-62
xla:380507 ppp3ca, MGC52521; protein phosphatase 3 (formerly 2... 234 4e-62
mmu:19057 Ppp3cc, Calnc; protein phosphatase 3, catalytic subu... 229 2e-60
sce:YML057W CMP2, CNA2; Calcineurin A; one isoform (the other ... 210 8e-55
sce:YLR433C CNA1, CMP1; Calcineurin A; one isoform (the other ... 205 2e-53
tgo:TGME49_111310 serine/threonine protein phosphatase, putati... 164 4e-41
pfa:PF08_0129 serine/threonine protein phosphatase, putative; ... 160 8e-40
cpv:cgd6_4200 protein phosphatase 2B catalytic subunit, calcin... 155 4e-38
ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphop... 61.2 7e-10
tgo:TGME49_112200 serine/threonine protein phosphatase, putati... 58.2 6e-09
pfa:MAL13P1.274 PfPP5; serine/threonine protein phosphatase (E... 52.8 3e-07
tpv:TP02_0944 serine/threonine protein phosphatase; K04460 pro... 52.8 3e-07
bbo:BBOV_IV000160 21.m02802; serine/threonine protein phosphat... 51.6 7e-07
hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein ph... 50.8 1e-06
cpv:cgd2_2960 phosphoprotein phosphatase related 50.1 2e-06
xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subuni... 50.1 2e-06
dre:541536 fc83f08, im:7146608, ppp5c, wu:fc83f08; zgc:110801 ... 49.3 3e-06
mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalyt... 48.9 4e-06
cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);... 47.8 8e-06
ath:AT4G11240 TOPP7; TOPP7; protein serine/threonine phosphatase 45.1 6e-05
cel:C05A2.1 pph-1; Protein PHosphatase family member (pph-1); ... 43.5 1e-04
ath:AT5G27840 TOPP8; TOPP8; protein serine/threonine phosphatase 43.5 2e-04
dre:100333823 protein phosphatase 1, catalytic subunit, gamma-... 43.5 2e-04
ath:AT2G39840 TOPP4; TOPP4; protein serine/threonine phosphata... 43.1 2e-04
ath:AT2G29400 TOPP1; TOPP1 (TYPE ONE PROTEIN PHOSPHATASE 1); p... 42.7 3e-04
mmu:19047 Ppp1cc, PP1, dis2m1; protein phosphatase 1, catalyti... 42.4 3e-04
ath:AT3G05580 serine/threonine protein phosphatase, putative; ... 42.4 3e-04
hsa:5501 PPP1CC, PP1gamma, PPP1G; protein phosphatase 1, catal... 42.4 3e-04
xla:380598 ppp1cc, MGC64327, PP-1G-B, ppp1cc-B, xPP1-zeta; pro... 42.4 3e-04
xla:397767 ppp1cc, MGC85040, ppp1cc-a, ppp1g; protein phosphat... 42.4 3e-04
hsa:5499 PPP1CA, MGC15877, MGC1674, PP-1A, PP1alpha, PPP1A; pr... 42.4 3e-04
tgo:TGME49_110700 serine/threonine protein phosphatase, putati... 42.4 3e-04
tpv:TP02_0042 serine/threonine protein phosphatase; K01090 pro... 42.4 3e-04
mmu:19045 Ppp1ca, Ppp1c, dism2; protein phosphatase 1, catalyt... 42.4 4e-04
ath:AT1G64040 TOPP3; TOPP3; protein serine/threonine phosphata... 42.4 4e-04
cel:F23B12.1 hypothetical protein; K01090 protein phosphatase ... 42.4 4e-04
ath:AT5G59160 TOPP2; TOPP2; protein serine/threonine phosphatase 42.4 4e-04
sce:YER133W GLC7, CID1, DIS2; Glc7p (EC:3.1.3.16); K06269 prot... 42.0 5e-04
dre:407980 ppp1caa, wu:fc04c08, wu:fc09b07, wu:fc30g11, wu:fe0... 41.6 6e-04
> dre:550554 ppp3cb, wu:fq19c08, zgc:110141; protein phosphatase
3, catalytic subunit, beta isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=511
Score = 243 bits (619), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 112/124 (90%), Positives = 118/124 (95%), Gaps = 0/124 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A CE LQ NNLLS+IRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 250 SYPAVCEFLQTNNLLSVIRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 309
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVL+ICSDDEL+N+
Sbjct 310 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLSICSDDELMND 369
Query 121 GEEI 124
G+EI
Sbjct 370 GDEI 373
> dre:794574 ppp3ca, fj46e08, si:dkeyp-79c2.2, wu:fj46e08; protein
phosphatase 3, catalytic subunit, alpha isozyme; K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 241 bits (616), Expect = 3e-64, Method: Compositional matrix adjust.
Identities = 112/124 (90%), Positives = 118/124 (95%), Gaps = 0/124 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A C+ LQ NNLLSIIRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 260 SYPAVCDFLQNNNLLSIIRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 319
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVL+ICSDDEL+N+
Sbjct 320 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLSICSDDELMND 379
Query 121 GEEI 124
G+EI
Sbjct 380 GDEI 383
> cel:C02F4.2 tax-6; abnormal CHEmotaxis family member (tax-6);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=535
Score = 239 bits (610), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 110/120 (91%), Positives = 114/120 (95%), Gaps = 0/120 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY ACC+ LQ NNLLSIIRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 286 SYAACCDFLQHNNLLSIIRAHEAQDAGYRMYRKSQATGFPSLITIFSAPNYLDVYNNKAA 345
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLV++LNICSDDEL+ E
Sbjct 346 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVHILNICSDDELMAE 405
> hsa:5532 PPP3CB, CALNA2, CALNB, CNA2, PP2Bbeta; protein phosphatase
3, catalytic subunit, beta isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=525
Score = 239 bits (610), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 111/122 (90%), Positives = 115/122 (94%), Gaps = 0/122 (0%)
Query 2 YGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAI 61
Y A CE LQ NNLLSIIRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA+
Sbjct 271 YPAVCEFLQNNNLLSIIRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAAV 330
Query 62 LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNEG 121
LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVL+ICSDDEL+ EG
Sbjct 331 LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLSICSDDELMTEG 390
Query 122 EE 123
E+
Sbjct 391 ED 392
> mmu:19056 Ppp3cb, 1110063J16Rik, Calnb, CnAbeta, Cnab; protein
phosphatase 3, catalytic subunit, beta isoform (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=525
Score = 239 bits (610), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 111/122 (90%), Positives = 115/122 (94%), Gaps = 0/122 (0%)
Query 2 YGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAI 61
Y A CE LQ NNLLSIIRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA+
Sbjct 271 YPAVCEFLQNNNLLSIIRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAAV 330
Query 62 LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNEG 121
LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVL+ICSDDEL+ EG
Sbjct 331 LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLSICSDDELMTEG 390
Query 122 EE 123
E+
Sbjct 391 ED 392
> xla:432106 ppp3cb, MGC81043, calna2, calnb, cna2, pp2bbeta;
protein phosphatase 3, catalytic subunit, beta isozyme; K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=507
Score = 238 bits (608), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 109/123 (88%), Positives = 116/123 (94%), Gaps = 0/123 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A CE LQ NNLLS+IRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 259 SYPAVCEFLQSNNLLSVIRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 318
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVN+L+ICSDDEL+ +
Sbjct 319 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNILSICSDDELITD 378
Query 121 GEE 123
GE+
Sbjct 379 GED 381
> hsa:5533 PPP3CC, CALNA3, CNA3, PP2Bgamma; protein phosphatase
3, catalytic subunit, gamma isozyme (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=512
Score = 238 bits (608), Expect = 3e-63, Method: Compositional matrix adjust.
Identities = 111/122 (90%), Positives = 115/122 (94%), Gaps = 0/122 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A CE LQ NNLLSIIRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 257 SYPAVCEFLQNNNLLSIIRAHEAQDAGYRMYRKSQATGFPSLITIFSAPNYLDVYNNKAA 316
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDEL+++
Sbjct 317 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELISD 376
Query 121 GE 122
E
Sbjct 377 DE 378
> dre:557926 ppp3cca, ppp3cc, sb:cb767, si:dkey-21k4.5; protein
phosphatase 3, catalytic subunit, gamma isozyme, a (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=499
Score = 238 bits (606), Expect = 5e-63, Method: Compositional matrix adjust.
Identities = 109/124 (87%), Positives = 117/124 (94%), Gaps = 0/124 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A C+ L NNLLS+IRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 259 SYAAVCDFLTNNNLLSVIRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 318
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDEL++E
Sbjct 319 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELMSE 378
Query 121 GEEI 124
G+++
Sbjct 379 GDDL 382
> hsa:5530 PPP3CA, CALN, CALNA, CALNA1, CCN1, CNA1, PPP2B; protein
phosphatase 3, catalytic subunit, alpha isozyme (EC:3.1.3.16);
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 237 bits (604), Expect = 8e-63, Method: Compositional matrix adjust.
Identities = 111/126 (88%), Positives = 115/126 (91%), Gaps = 0/126 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A CE LQ NNLLSI+RAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 261 SYPAVCEFLQHNNLLSILRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 320
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDEL +E
Sbjct 321 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELGSE 380
Query 121 GEEIDN 126
+ D
Sbjct 381 EDGFDG 386
> dre:565241 ppp3ccb, im:7163784, zgc:158685; protein phosphatase
3, catalytic subunit, gamma isozyme, b (EC:3.1.3.16); K04348
protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=506
Score = 236 bits (602), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 109/123 (88%), Positives = 116/123 (94%), Gaps = 0/123 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A C+ L NNLLS+IRAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 258 SYPAVCDFLTNNNLLSVIRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 317
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDEL++E
Sbjct 318 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELMSE 377
Query 121 GEE 123
G++
Sbjct 378 GDD 380
> xla:447196 ppp3ca, MGC79144, calcineurin, caln, calna, calna1,
ccn1, cna1, ppp2b; protein phosphatase 3, catalytic subunit,
alpha isozyme; K04348 protein phosphatase 3, catalytic subunit
[EC:3.1.3.16]
Length=511
Score = 236 bits (602), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 109/126 (86%), Positives = 116/126 (92%), Gaps = 0/126 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A CE LQ NNLLS++RAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 261 SYPAVCEFLQHNNLLSVLRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 320
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVL+ICSDDEL++E
Sbjct 321 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLSICSDDELVSE 380
Query 121 GEEIDN 126
+ D
Sbjct 381 EDACDG 386
> mmu:19055 Ppp3ca, 2900074D19Rik, AI841391, AW413465, CN, Caln,
Calna, CnA, MGC106804; protein phosphatase 3, catalytic subunit,
alpha isoform (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=521
Score = 236 bits (601), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 110/126 (87%), Positives = 115/126 (91%), Gaps = 0/126 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A C+ LQ NNLLSI+RAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 261 SYPAVCDFLQHNNLLSILRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 320
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDEL +E
Sbjct 321 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELGSE 380
Query 121 GEEIDN 126
+ D
Sbjct 381 EDGFDG 386
> xla:380507 ppp3ca, MGC52521; protein phosphatase 3 (formerly
2B), catalytic subunit, alpha isoform (calcineurin A alpha)
(EC:3.1.3.16); K04348 protein phosphatase 3, catalytic subunit
[EC:3.1.3.16]
Length=518
Score = 234 bits (598), Expect = 4e-62, Method: Compositional matrix adjust.
Identities = 108/126 (85%), Positives = 116/126 (92%), Gaps = 0/126 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A CE LQ NNLLS++RAHEAQDAGYRMYRKS TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 261 SYPAVCEFLQHNNLLSVLRAHEAQDAGYRMYRKSQTTGFPSLITIFSAPNYLDVYNNKAA 320
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVL+ICSDDEL+++
Sbjct 321 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLSICSDDELVSD 380
Query 121 GEEIDN 126
+ D
Sbjct 381 EDACDG 386
> mmu:19057 Ppp3cc, Calnc; protein phosphatase 3, catalytic subunit,
gamma isoform (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=513
Score = 229 bits (583), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 105/117 (89%), Positives = 111/117 (94%), Gaps = 0/117 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
SY A CE LQ N+LLSIIRAHEAQDAGYRMYRK+ TGFPSLITIFSAPNYLDVYNNKAA
Sbjct 257 SYPAVCEFLQNNSLLSIIRAHEAQDAGYRMYRKNQATGFPSLITIFSAPNYLDVYNNKAA 316
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDEL 117
+LKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVN+LNICSD+E+
Sbjct 317 VLKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNILNICSDEEM 373
> sce:YML057W CMP2, CNA2; Calcineurin A; one isoform (the other
is CNA1) of the catalytic subunit of calcineurin, a Ca++/calmodulin-regulated
protein phosphatase which regulates Crz1p
(a stress-response transcription factor), the other calcineurin
subunit is CNB1 (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=604
Score = 210 bits (535), Expect = 8e-55, Method: Compositional matrix adjust.
Identities = 95/125 (76%), Positives = 108/125 (86%), Gaps = 0/125 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
+Y A C LQ+ LLSIIRAHEAQDAGYRMY+ + GFPSL+T+FSAPNYLD YNNKAA
Sbjct 339 TYRAACHFLQETGLLSIIRAHEAQDAGYRMYKNTKTLGFPSLLTLFSAPNYLDTYNNKAA 398
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
ILKYENNVMNIRQFN +PHPYWLP+FMDVFTWSLPFVGEKVTEMLV +LNIC++DEL N+
Sbjct 399 ILKYENNVMNIRQFNMTPHPYWLPDFMDVFTWSLPFVGEKVTEMLVAILNICTEDELEND 458
Query 121 GEEID 125
I+
Sbjct 459 TPVIE 463
> sce:YLR433C CNA1, CMP1; Calcineurin A; one isoform (the other
is CMP2) of the catalytic subunit of calcineurin, a Ca++/calmodulin-regulated
protein phosphatase which regulates Crz1p
(a stress-response transcription factor), the other calcineurin
subunit is CNB1 (EC:3.1.3.16); K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=553
Score = 205 bits (522), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 93/122 (76%), Positives = 107/122 (87%), Gaps = 0/122 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
++ A C+ L+ N LLSIIRAHEAQDAGYRMY+ + VTGFPSLIT+FSAPNYLD Y+NKAA
Sbjct 297 TFKASCKFLKANGLLSIIRAHEAQDAGYRMYKNNKVTGFPSLITMFSAPNYLDTYHNKAA 356
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDDELLNE 120
+LKYE NVMNIRQF+ SPHPYWLP+FMDVFTWSLPFVGEKVT MLV++LNICS+ EL E
Sbjct 357 VLKYEENVMNIRQFHMSPHPYWLPDFMDVFTWSLPFVGEKVTSMLVSILNICSEQELDPE 416
Query 121 GE 122
E
Sbjct 417 SE 418
> tgo:TGME49_111310 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04348 protein phosphatase 3, catalytic
subunit [EC:3.1.3.16]
Length=501
Score = 164 bits (416), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 74/110 (67%), Positives = 90/110 (81%), Gaps = 0/110 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
Y A + L +N LLS++RAHEAQ GY+M++ + TGFP++ITIFSAPNY DVYNNK A
Sbjct 261 GYSAASKFLDRNGLLSVLRAHEAQLEGYKMHQTNQKTGFPTVITIFSAPNYCDVYNNKGA 320
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLN 110
+LK+ENN +NI+QFN SPHPY LPNFMDVFTWS+PFV EKVTEML +LN
Sbjct 321 VLKFENNTLNIQQFNFSPHPYHLPNFMDVFTWSIPFVSEKVTEMLYGILN 370
> pfa:PF08_0129 serine/threonine protein phosphatase, putative;
K04348 protein phosphatase 3, catalytic subunit [EC:3.1.3.16]
Length=604
Score = 160 bits (405), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 74/115 (64%), Positives = 93/115 (80%), Gaps = 0/115 (0%)
Query 1 SYGACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAA 60
Y A L++N LLSIIRAHEAQ GY+M++ +L TGFP +ITIFSAPNY DVYNNK A
Sbjct 260 GYNAATTFLEKNGLLSIIRAHEAQLEGYKMHQTNLKTGFPIVITIFSAPNYCDVYNNKGA 319
Query 61 ILKYENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLNICSDD 115
+LK+++N +NI+QF+ SPHPY LPNFM++FTWSLPFV EKVTEML ++LN +D
Sbjct 320 VLKFDSNTLNIQQFSFSPHPYHLPNFMNLFTWSLPFVSEKVTEMLYSLLNCSMND 374
> cpv:cgd6_4200 protein phosphatase 2B catalytic subunit, calcineurin
like phosphatse superfamily ; K04348 protein phosphatase
3, catalytic subunit [EC:3.1.3.16]
Length=504
Score = 155 bits (391), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 70/107 (65%), Positives = 88/107 (82%), Gaps = 0/107 (0%)
Query 4 ACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILK 63
A + L QN LLSI+RAHEAQ G++M+ ++ T FPS+ITIFSAPNY DVYNNK AILK
Sbjct 262 ASNKFLDQNGLLSIVRAHEAQLEGFKMHSPNVKTRFPSVITIFSAPNYCDVYNNKGAILK 321
Query 64 YENNVMNIRQFNCSPHPYWLPNFMDVFTWSLPFVGEKVTEMLVNVLN 110
+ENN +NI+QFN S HPY+LPNF+D+F+WSLPFV EKVTEML +++
Sbjct 322 FENNTLNIQQFNFSAHPYYLPNFLDIFSWSLPFVSEKVTEMLYSIIQ 368
> ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphoprotein
phosphatase/ protein binding / protein serine/threonine
phosphatase; K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=538
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 36/91 (39%), Positives = 48/91 (52%), Gaps = 13/91 (14%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
LQ NNL ++R+HE +D GY + LIT+FSAPNY D NK A +++E
Sbjct 453 RFLQDNNLDLLVRSHEVKDEGYEVEHDG------KLITVFSAPNYCDQMGNKGAFIRFEA 506
Query 67 NVM--NIRQFNCSPHPYWLP-----NFMDVF 90
M NI F+ PHP P NF+ +F
Sbjct 507 PDMKPNIVTFSAVPHPDVKPMAYANNFLRMF 537
> tgo:TGME49_112200 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=548
Score = 58.2 bits (139), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 42/79 (53%), Gaps = 8/79 (10%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENN 67
L++NNL +IR+HE +D GY + L+T+FSAPNY D NK A ++ +
Sbjct 465 FLKRNNLKLVIRSHEMKDDGYEVCHNG------QLVTVFSAPNYCDQMKNKGAFVRLDGK 518
Query 68 VMNIR--QFNCSPHPYWLP 84
+ + QF PHP P
Sbjct 519 TLTPKYTQFEAVPHPATRP 537
> pfa:MAL13P1.274 PfPP5; serine/threonine protein phosphatase
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=658
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 37/62 (59%), Gaps = 6/62 (9%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENN 67
L+ NNL IIR+HE +D GY + + L T+FSAPNY D+ NK A LK++ N
Sbjct 575 FLKINNLSLIIRSHEVRDEGYSLEQNG------QLYTVFSAPNYCDIMKNKGAFLKFKGN 628
Query 68 VM 69
+
Sbjct 629 SI 630
> tpv:TP02_0944 serine/threonine protein phosphatase; K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=548
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 41/75 (54%), Gaps = 8/75 (10%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENN 67
+ +NNL IIR+HE + GY + + V ITIFSAPNY D NKAA ++ + +
Sbjct 464 FISKNNLSYIIRSHEVKQEGYEVEHEGKV------ITIFSAPNYCDQMGNKAAFIRIKGD 517
Query 68 VMNIR--QFNCSPHP 80
+ + QF HP
Sbjct 518 TLEPKFTQFEAVEHP 532
> bbo:BBOV_IV000160 21.m02802; serine/threonine protein phosphatase
5 (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=545
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 30/75 (40%), Positives = 38/75 (50%), Gaps = 8/75 (10%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENN 67
LQQN L IIR+HE + GY + LITIFSAPNY D NK A ++ +
Sbjct 461 FLQQNGLELIIRSHEVKQEGYELEHNG------QLITIFSAPNYCDQMGNKGAFIRMNGS 514
Query 68 VMNIR--QFNCSPHP 80
+ QF+ HP
Sbjct 515 DCKPKFTQFDAVEHP 529
> hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein
phosphatase 5, catalytic subunit (EC:3.1.3.16); K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=477
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 40/79 (50%), Gaps = 8/79 (10%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENN 67
L++NNL IIR+HE + GY V +T+FSAPNY D NKA+ + + +
Sbjct 392 FLEENNLDYIIRSHEVKAEGYE------VAHGGRCVTVFSAPNYCDQMGNKASYIHLQGS 445
Query 68 VM--NIRQFNCSPHPYWLP 84
+ QF PHP P
Sbjct 446 DLRPQFHQFTAVPHPNVKP 464
> cpv:cgd2_2960 phosphoprotein phosphatase related
Length=525
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/75 (41%), Positives = 40/75 (53%), Gaps = 8/75 (10%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILK-YEN 66
L+ NNL IIR+HE + GY + IT+FSAPNY D NK A +K +E
Sbjct 441 FLKTNNLDYIIRSHEVKQEGYSVDHDG------KCITVFSAPNYCDSMGNKGAFIKIHEY 494
Query 67 NVM-NIRQFNCSPHP 80
++ N QF PHP
Sbjct 495 DLKPNFVQFYAVPHP 509
> xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subunit
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=493
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 39/80 (48%), Gaps = 8/80 (10%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
+ L++N L IIR+HE + GY + L +T+FSAPNY D NK A +
Sbjct 407 QFLEENGLDYIIRSHEVKPEGYEVSHNGLC------VTVFSAPNYCDQMGNKGAYIHLSG 460
Query 67 NVM--NIRQFNCSPHPYWLP 84
+ + QF PHP P
Sbjct 461 SDLKPKFHQFEAVPHPNVKP 480
> dre:541536 fc83f08, im:7146608, ppp5c, wu:fc83f08; zgc:110801
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=481
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 37/79 (46%), Gaps = 8/79 (10%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENN 67
L+QN L I+R+HE GY VT IT+FSAPNY D NK A + +
Sbjct 396 FLEQNKLEYIVRSHEVIAEGYE------VTHSGKCITVFSAPNYCDQMGNKEAYIHLRGS 449
Query 68 VMN--IRQFNCSPHPYWLP 84
+ QF PHP P
Sbjct 450 DLKPEFHQFTAVPHPNVKP 468
> mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalytic
subunit (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=499
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 39/79 (49%), Gaps = 8/79 (10%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENN 67
L++N L IIR+HE + GY V +T+FSAPNY D NKA+ + + +
Sbjct 414 FLEENQLDYIIRSHEVKAEGYE------VAHGGRCVTVFSAPNYCDQMGNKASYIHLQGS 467
Query 68 VM--NIRQFNCSPHPYWLP 84
+ QF PHP P
Sbjct 468 DLRPQFHQFTAVPHPNVKP 486
> cel:Y39B6A.2 pph-5; Protein PHosphatase family member (pph-5);
K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=496
Score = 47.8 bits (112), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 37/77 (48%), Gaps = 8/77 (10%)
Query 10 QQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENNVM 69
+ N + ++R+HE + GY M+ T+FSAPNY D NNK A + + +
Sbjct 415 ETNGIEYVVRSHEVKPEGYEMHHNG------QCFTVFSAPNYCDQMNNKGAFITITGDNL 468
Query 70 NIR--QFNCSPHPYWLP 84
R F+ PHP P
Sbjct 469 TPRFTPFDAVPHPKLPP 485
> ath:AT4G11240 TOPP7; TOPP7; protein serine/threonine phosphatase
Length=322
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 40/70 (57%), Gaps = 8/70 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA E LQ ++L I RAH+ + GY + K L+TIFSAPNY ++N
Sbjct 223 TFGADKVAEFLQTHDLDLICRAHQVVEDGYEFFAKR------QLVTIFSAPNYCGEFDNA 276
Query 59 AAILKYENNV 68
A++ ++++
Sbjct 277 GALMSVDDSL 286
> cel:C05A2.1 pph-1; Protein PHosphatase family member (pph-1);
K01090 protein phosphatase [EC:3.1.3.16]
Length=349
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 36/63 (57%), Gaps = 6/63 (9%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
+ LQ+ +L ++R H+ + GY + G L+T+FSAPNY ++N A++ +
Sbjct 239 QFLQKMDLDIVVRGHQVVEDGYEFF------GRRGLVTVFSAPNYCGEFDNAGAVMNVDE 292
Query 67 NVM 69
N++
Sbjct 293 NLL 295
> ath:AT5G27840 TOPP8; TOPP8; protein serine/threonine phosphatase
Length=324
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA E L +N+L I R H+ + GY + K L+TIFSAPNY ++N
Sbjct 228 TFGADKVAEFLDKNDLDLICRGHQVVEDGYEFFAKR------RLVTIFSAPNYGGEFDNA 281
Query 59 AAILKYENNVM 69
A+L + +++
Sbjct 282 GALLSVDESLV 292
> dre:100333823 protein phosphatase 1, catalytic subunit, gamma-like
Length=158
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA + L +++L I RAH+ + GY + K L+T+FSAPNY ++N
Sbjct 61 TFGAEVVAKFLHKHDLDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNA 114
Query 59 AAILKYENNVM 69
A++ + +M
Sbjct 115 GAMMSVDETLM 125
> ath:AT2G39840 TOPP4; TOPP4; protein serine/threonine phosphatase;
K06269 protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=321
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 35/63 (55%), Gaps = 6/63 (9%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
E L +++L + RAH+ + GY + L+T+FSAPNY ++N A++ +
Sbjct 244 EFLTKHDLDLVCRAHQVVEDGYEFFADR------QLVTVFSAPNYCGEFDNAGAMMSVDE 297
Query 67 NVM 69
N+M
Sbjct 298 NLM 300
> ath:AT2G29400 TOPP1; TOPP1 (TYPE ONE PROTEIN PHOSPHATASE 1);
protein serine/threonine phosphatase
Length=318
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 41/71 (57%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA E L+++++ I RAH+ + GY + + L+T+FSAPNY ++N
Sbjct 238 TFGADKVAEFLEKHDMDLICRAHQVVEDGYEFFAER------QLVTVFSAPNYCGEFDNA 291
Query 59 AAILKYENNVM 69
A++ + ++M
Sbjct 292 GAMMSIDESLM 302
> mmu:19047 Ppp1cc, PP1, dis2m1; protein phosphatase 1, catalytic
subunit, gamma isoform (EC:3.1.3.16); K06269 protein phosphatase
1, catalytic subunit [EC:3.1.3.16]
Length=323
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA + L +++L I RAH+ + GY + K L+T+FSAPNY ++N
Sbjct 226 TFGAEVVAKFLHKHDLDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNA 279
Query 59 AAILKYENNVM 69
A++ + +M
Sbjct 280 GAMMSVDETLM 290
> ath:AT3G05580 serine/threonine protein phosphatase, putative;
K01090 protein phosphatase [EC:3.1.3.16]
Length=318
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA + L +N+L I R H+ + GY + K L+TIFSAPNY ++N
Sbjct 228 TFGADVVADFLDKNDLDLICRGHQVVEDGYEFFAKR------RLVTIFSAPNYGGEFDNA 281
Query 59 AAILKYENNVM 69
A+L + +++
Sbjct 282 GALLSVDQSLV 292
> hsa:5501 PPP1CC, PP1gamma, PPP1G; protein phosphatase 1, catalytic
subunit, gamma isozyme (EC:3.1.3.16); K06269 protein
phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=323
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA + L +++L I RAH+ + GY + K L+T+FSAPNY ++N
Sbjct 226 TFGAEVVAKFLHKHDLDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNA 279
Query 59 AAILKYENNVM 69
A++ + +M
Sbjct 280 GAMMSVDETLM 290
> xla:380598 ppp1cc, MGC64327, PP-1G-B, ppp1cc-B, xPP1-zeta; protein
phosphatase 1, catalytic subunit, gamma isoform (EC:3.1.3.16);
K06269 protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=323
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA + L +++L I RAH+ + GY + K L+T+FSAPNY ++N
Sbjct 226 TFGAEVVAKFLHKHDLDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNA 279
Query 59 AAILKYENNVM 69
A++ + +M
Sbjct 280 GAMMSVDETLM 290
> xla:397767 ppp1cc, MGC85040, ppp1cc-a, ppp1g; protein phosphatase
1, catalytic subunit, gamma isozyme (EC:3.1.3.16); K06269
protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=323
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA + L +++L I RAH+ + GY + K L+T+FSAPNY ++N
Sbjct 226 TFGAEVVAKFLHKHDLDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNA 279
Query 59 AAILKYENNVM 69
A++ + +M
Sbjct 280 GAMMSVDETLM 290
> hsa:5499 PPP1CA, MGC15877, MGC1674, PP-1A, PP1alpha, PPP1A;
protein phosphatase 1, catalytic subunit, alpha isozyme (EC:3.1.3.16);
K06269 protein phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=341
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 35/63 (55%), Gaps = 6/63 (9%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
+ L +++L I RAH+ + GY + K L+T+FSAPNY ++N A++ +
Sbjct 245 KFLHKHDLDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNAGAMMSVDE 298
Query 67 NVM 69
+M
Sbjct 299 TLM 301
> tgo:TGME49_110700 serine/threonine protein phosphatase, putative
(EC:3.1.3.16); K06269 protein phosphatase 1, catalytic
subunit [EC:3.1.3.16]
Length=306
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 35/62 (56%), Gaps = 6/62 (9%)
Query 8 LLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYENN 67
L++++L I RAH+ + GY + K L+T+FSAPNY ++N A++ +
Sbjct 234 FLRKHDLDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNAGAMMSVDET 287
Query 68 VM 69
+M
Sbjct 288 LM 289
> tpv:TP02_0042 serine/threonine protein phosphatase; K01090 protein
phosphatase [EC:3.1.3.16]
Length=346
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 40/73 (54%), Gaps = 7/73 (9%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
+ QNN+ +I RAH+ GY+ + SLIT++SAPNY N A+I++ +
Sbjct 263 QFCHQNNIQTIARAHQLVMEGYKWWFNK------SLITVWSAPNYCYRCGNIASIMELDE 316
Query 67 NV-MNIRQFNCSP 78
N+ N ++F P
Sbjct 317 NLNCNFKKFEAVP 329
> mmu:19045 Ppp1ca, Ppp1c, dism2; protein phosphatase 1, catalytic
subunit, alpha isoform (EC:3.1.3.16); K06269 protein phosphatase
1, catalytic subunit [EC:3.1.3.16]
Length=330
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA + L +++L I RAH+ + GY + K L+T+FSAPNY ++N
Sbjct 226 TFGAEVVAKFLHKHDLDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNA 279
Query 59 AAILKYENNVM 69
A++ + +M
Sbjct 280 GAMMSVDETLM 290
> ath:AT1G64040 TOPP3; TOPP3; protein serine/threonine phosphatase;
K01090 protein phosphatase [EC:3.1.3.16]
Length=322
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 35/62 (56%), Gaps = 6/62 (9%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
E LQ ++L I RAH+ + GY + L+TIFSAPNY ++N A++ ++
Sbjct 231 EFLQTHDLDLICRAHQVVEDGYEFFANR------QLVTIFSAPNYCGEFDNAGAMMSVDD 284
Query 67 NV 68
++
Sbjct 285 SL 286
> cel:F23B12.1 hypothetical protein; K01090 protein phosphatase
[EC:3.1.3.16]
Length=401
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 40/71 (56%), Gaps = 8/71 (11%)
Query 1 SYG--ACCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
S+G A + LQ +NL ++RAH+ GY + L+T+FSAP+Y ++N
Sbjct 293 SFGPNAVKKFLQMHNLDLVVRAHQVVMDGYEFFADR------QLVTVFSAPSYCGQFDNA 346
Query 59 AAILKYENNVM 69
AA++ ++ ++
Sbjct 347 AAVMNVDDKLL 357
> ath:AT5G59160 TOPP2; TOPP2; protein serine/threonine phosphatase
Length=312
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 35/63 (55%), Gaps = 6/63 (9%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
E L +N++ I RAH+ + GY + L+TIFSAPNY ++N A++ +
Sbjct 240 EFLIKNDMDLICRAHQVVEDGYEFFADR------QLVTIFSAPNYCGEFDNAGAMMSVDE 293
Query 67 NVM 69
++M
Sbjct 294 SLM 296
> sce:YER133W GLC7, CID1, DIS2; Glc7p (EC:3.1.3.16); K06269 protein
phosphatase 1, catalytic subunit [EC:3.1.3.16]
Length=312
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 35/63 (55%), Gaps = 6/63 (9%)
Query 7 ELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNKAAILKYEN 66
LQ+ ++ I RAH+ + GY + K L+T+FSAPNY ++N A++ +
Sbjct 233 RFLQKQDMELICRAHQVVEDGYEFFSKR------QLVTLFSAPNYCGEFDNAGAMMSVDE 286
Query 67 NVM 69
+++
Sbjct 287 SLL 289
> dre:407980 ppp1caa, wu:fc04c08, wu:fc09b07, wu:fc30g11, wu:fe05h08,
zgc:85729; protein phosphatase 1, catalytic subunit,
alpha isoform a (EC:3.1.3.16); K06269 protein phosphatase 1,
catalytic subunit [EC:3.1.3.16]
Length=331
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 39/71 (54%), Gaps = 8/71 (11%)
Query 1 SYGA--CCELLQQNNLLSIIRAHEAQDAGYRMYRKSLVTGFPSLITIFSAPNYLDVYNNK 58
++GA + L ++++ I RAH+ + GY + K L+T+FSAPNY ++N
Sbjct 226 TFGADVVAKFLHKHDMDLICRAHQVVEDGYEFFAKR------QLVTLFSAPNYCGEFDNA 279
Query 59 AAILKYENNVM 69
A++ + +M
Sbjct 280 GAMMSVDETLM 290
Lambda K H
0.320 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2054672932
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40