bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2428_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
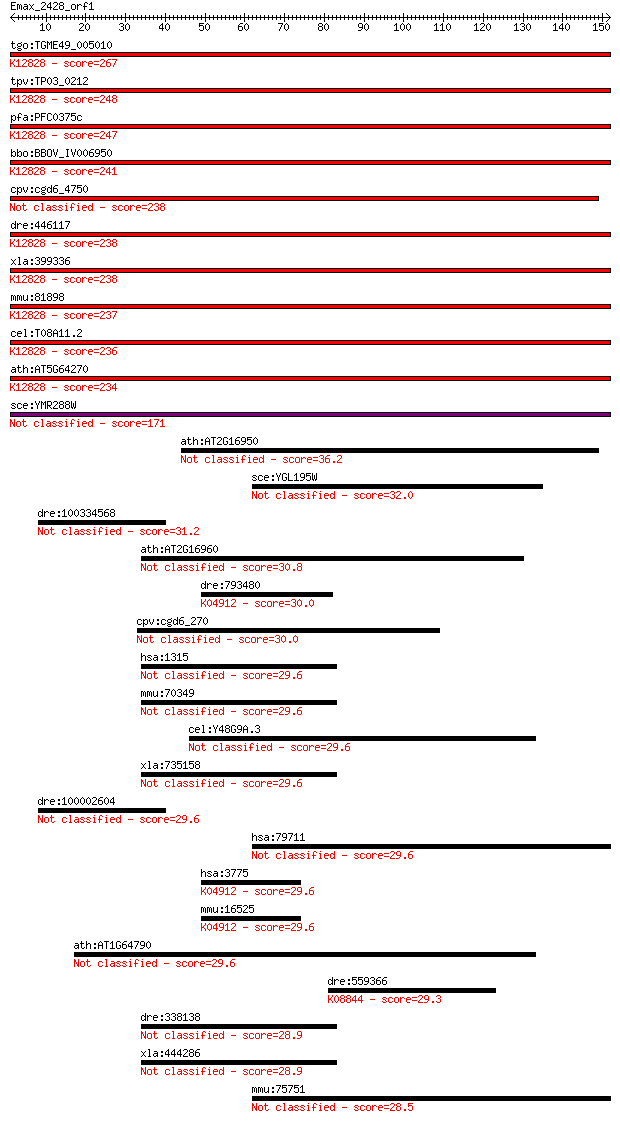
tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5... 267 8e-72
tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing fa... 248 6e-66
pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828 sp... 247 9e-66
bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing ... 241 5e-64
cpv:cgd6_4750 splicing factor 3B subunit1-like HEAT repeat con... 238 4e-63
dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1; K... 238 7e-63
xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155; sp... 238 8e-63
mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP15... 237 1e-62
cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B s... 236 2e-62
ath:AT5G64270 splicing factor, putative; K12828 splicing facto... 234 5e-62
sce:YMR288W HSH155; Hsh155p 171 7e-43
ath:AT2G16950 TRN1; TRN1 (TRANSPORTIN 1); protein transporter 36.2 0.038
sce:YGL195W GCN1, AAS103, NDR1; Gcn1p 32.0 0.79
dre:100334568 phosphodiesterase 5A-like 31.2 1.4
ath:AT2G16960 importin beta-2 subunit family protein 30.8 1.6
dre:793480 kcnk1, MGC165664, im:7150627, zgc:165664; potassium... 30.0 2.9
cpv:cgd6_270 hypothetical protein 30.0 3.2
hsa:1315 COPB1, COPB, DKFZp761K102, FLJ10341, FLJ46444, FLJ579... 29.6 3.4
mmu:70349 Copb1, 2610019B04Rik; coatomer protein complex, subu... 29.6 3.5
cel:Y48G9A.3 hypothetical protein 29.6 3.6
xla:735158 hypothetical protein MGC114704 29.6 3.7
dre:100002604 phosphodiesterase 5A-like 29.6 3.9
hsa:79711 IPO4, FLJ23338, Imp4, MGC131665; importin 4 29.6
hsa:3775 KCNK1, DPK, HOHO, K2P1, K2p1.1, KCNO1, TWIK-1, TWIK1;... 29.6 4.2
mmu:16525 Kcnk1, AI788889, TWIK-1; potassium channel, subfamil... 29.6 4.2
ath:AT1G64790 binding 29.6 4.4
dre:559366 lrrk2; leucine-rich repeat kinase 2 (EC:2.7.11.1); ... 29.3 4.4
dre:338138 copb1, MGC92482, id:ibd5034, wu:fc66c05, zgc:92482;... 28.9 6.5
xla:444286 copb1, MGC80934; coatomer protein complex, subunit ... 28.9 6.9
mmu:75751 Ipo4, 8430408O15Rik, AA409693, Imp4a, MGC113723, Ran... 28.5 7.7
> tgo:TGME49_005010 splicing factor 3B subunit 1, putative (EC:5.5.1.4);
K12828 splicing factor 3B subunit 1
Length=1386
Score = 267 bits (683), Expect = 8e-72, Method: Compositional matrix adjust.
Identities = 138/151 (91%), Positives = 147/151 (97%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVV SALG
Sbjct 662 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVASALG 721
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+PSL+LFLKAVCQSKKSWQARHTGI+IIQQ++IL+GCGVLPHL+Q VEIIQHGLDD VLK
Sbjct 722 IPSLILFLKAVCQSKKSWQARHTGIKIIQQMSILMGCGVLPHLKQLVEIIQHGLDDQVLK 781
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
+KTVTALALAALAE+AAPYGIEAFD VLRPL
Sbjct 782 VKTVTALALAALAESAAPYGIEAFDSVLRPL 812
> tpv:TP03_0212 splicing factor 3B subunit 1; K12828 splicing
factor 3B subunit 1
Length=1107
Score = 248 bits (633), Expect = 6e-66, Method: Compositional matrix adjust.
Identities = 122/151 (80%), Positives = 142/151 (94%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNL+KAAGLATMI MRPDIDHPDEYVRNTTARAFAVV S++G
Sbjct 372 LLIDEDYYARVEGREIISNLSKAAGLATMIGVMRPDIDHPDEYVRNTTARAFAVVASSMG 431
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
V SL+LFLKAVCQSKKSWQARHTGI+IIQQI++L+GCGVLP+L+Q ++II+HGL+D K
Sbjct 432 VSSLILFLKAVCQSKKSWQARHTGIKIIQQISLLIGCGVLPYLKQLIDIIKHGLNDEHQK 491
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T+TALALAALAE++APYGIEAFD VLRPL
Sbjct 492 VRTITALALAALAESSAPYGIEAFDPVLRPL 522
> pfa:PFC0375c U2 snRNP spliceosome subunit, putative; K12828
splicing factor 3B subunit 1
Length=1386
Score = 247 bits (631), Expect = 9e-66, Method: Composition-based stats.
Identities = 125/151 (82%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNLAKAAGLATMI MRPDIDHPDEYVRNTTARAFAVV SALG
Sbjct 662 LLIDEDYYARVEGREIISNLAKAAGLATMIGIMRPDIDHPDEYVRNTTARAFAVVASALG 721
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+PSL+LFLKAVCQSKK+W+ARHTGI+I+QQIAIL+GC VLPHL+ V+II HGL D K
Sbjct 722 IPSLILFLKAVCQSKKNWEARHTGIKIVQQIAILMGCAVLPHLKDLVQIIAHGLHDEQQK 781
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T+TALA+AALAEAAAPYGIEAFD VLRPL
Sbjct 782 VRTITALAVAALAEAAAPYGIEAFDSVLRPL 812
> bbo:BBOV_IV006950 23.m06336; splicing factor; K12828 splicing
factor 3B subunit 1
Length=1147
Score = 241 bits (616), Expect = 5e-64, Method: Compositional matrix adjust.
Identities = 125/151 (82%), Positives = 140/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNL+KAAGLATMI TMRPDIDHPDEYVRNTTARAFAVV A G
Sbjct 418 LLIDEDYYARVEGREIISNLSKAAGLATMIGTMRPDIDHPDEYVRNTTARAFAVVAHATG 477
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+ SL+LFLKAVCQSKKSWQARHTGI+I+QQIAIL+GCGVLPHL+Q V II GL+D VLK
Sbjct 478 IQSLILFLKAVCQSKKSWQARHTGIKIVQQIAILVGCGVLPHLKQLVSIIASGLEDEVLK 537
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T+TALALA+LAEA+AP+GIEAFD VLRPL
Sbjct 538 VRTMTALALASLAEASAPFGIEAFDIVLRPL 568
> cpv:cgd6_4750 splicing factor 3B subunit1-like HEAT repeat containing
protein
Length=1031
Score = 238 bits (608), Expect = 4e-63, Method: Compositional matrix adjust.
Identities = 110/148 (74%), Positives = 132/148 (89%), Gaps = 0/148 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
+LID+DYYAR EGREIISNLAKA GLATMIATMRPDIDHPDEYVRNTTA+AFA++ SA+G
Sbjct 307 MLIDQDYYARQEGREIISNLAKAVGLATMIATMRPDIDHPDEYVRNTTAKAFAILASAMG 366
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+PSL++FL+AVCQSKKSWQARHTGI I+QQIAIL G VLPHL+ V+II HGL D K
Sbjct 367 IPSLVIFLQAVCQSKKSWQARHTGIRIVQQIAILHGSSVLPHLKSLVQIISHGLSDENQK 426
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVL 148
++ +TAL+LA+LAEA++PYGIEAF+ +L
Sbjct 427 VRVITALSLASLAEASSPYGIEAFEPIL 454
> dre:446117 sf3b1, wu:fb99f09; splicing factor 3b, subunit 1;
K12828 splicing factor 3B subunit 1
Length=1315
Score = 238 bits (606), Expect = 7e-63, Method: Compositional matrix adjust.
Identities = 122/151 (80%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNLAKAAGLATMI+TMRPDID+ DEYVRNTTARAFAVV SALG
Sbjct 592 LLIDEDYYARVEGREIISNLAKAAGLATMISTMRPDIDNMDEYVRNTTARAFAVVASALG 651
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+PSLL FLKAVC+SKKSWQARHTGI+I+QQIAIL+GC +LPHLR VEII+HGL D K
Sbjct 652 IPSLLPFLKAVCKSKKSWQARHTGIKIVQQIAILMGCAILPHLRSLVEIIEHGLVDEQQK 711
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T++ALA+AALAEAA PYGIE+FD VL+PL
Sbjct 712 VRTISALAIAALAEAATPYGIESFDSVLKPL 742
> xla:399336 sf3b1, MGC115390, hsh155, prp10, prpf10, sap155;
splicing factor 3b, subunit 1, 155kDa; K12828 splicing factor
3B subunit 1
Length=1307
Score = 238 bits (606), Expect = 8e-63, Method: Compositional matrix adjust.
Identities = 122/151 (80%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNLAKAAGLATMI+TMRPDID+ DEYVRNTTARAFAVV SALG
Sbjct 584 LLIDEDYYARVEGREIISNLAKAAGLATMISTMRPDIDNMDEYVRNTTARAFAVVASALG 643
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+PSLL FLKAVC+SKKSWQARHTGI+I+QQIAIL+GC +LPHLR VEII+HGL D K
Sbjct 644 IPSLLPFLKAVCKSKKSWQARHTGIKIVQQIAILMGCAILPHLRSLVEIIEHGLVDEQQK 703
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T++ALA+AALAEAA PYGIE+FD VL+PL
Sbjct 704 VRTISALAIAALAEAATPYGIESFDSVLKPL 734
> mmu:81898 Sf3b1, 155kDa, 2810001M05Rik, AA409119, Prp10, SAP155,
SF3b155, TA-8, Targ4; splicing factor 3b, subunit 1; K12828
splicing factor 3B subunit 1
Length=1304
Score = 237 bits (604), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 122/151 (80%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNLAKAAGLATMI+TMRPDID+ DEYVRNTTARAFAVV SALG
Sbjct 581 LLIDEDYYARVEGREIISNLAKAAGLATMISTMRPDIDNMDEYVRNTTARAFAVVASALG 640
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+PSLL FLKAVC+SKKSWQARHTGI+I+QQIAIL+GC +LPHLR VEII+HGL D K
Sbjct 641 IPSLLPFLKAVCKSKKSWQARHTGIKIVQQIAILMGCAILPHLRSLVEIIEHGLVDEQQK 700
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T++ALA+AALAEAA PYGIE+FD VL+PL
Sbjct 701 VRTISALAIAALAEAATPYGIESFDSVLKPL 731
> cel:T08A11.2 hypothetical protein; K12828 splicing factor 3B
subunit 1
Length=1322
Score = 236 bits (602), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 119/151 (78%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNLAKAAGLATMI+TMRPDID+ DEYVRNTTARAFAVV SALG
Sbjct 599 LLIDEDYYARVEGREIISNLAKAAGLATMISTMRPDIDNVDEYVRNTTARAFAVVASALG 658
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+P+LL FLKAVC+SKKSWQARHTGI+I+QQ+AIL+GC VLPHL+ V+I++ GLDD K
Sbjct 659 IPALLPFLKAVCKSKKSWQARHTGIKIVQQMAILMGCAVLPHLKALVDIVESGLDDEQQK 718
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T+TAL LAALAEA++PYGIEAFD VL+PL
Sbjct 719 VRTITALCLAALAEASSPYGIEAFDSVLKPL 749
> ath:AT5G64270 splicing factor, putative; K12828 splicing factor
3B subunit 1
Length=1269
Score = 234 bits (598), Expect = 5e-62, Method: Compositional matrix adjust.
Identities = 121/151 (80%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNL+KAAGLA+MIA MRPDID+ DEYVRNTTARAF+VV SALG
Sbjct 545 LLIDEDYYARVEGREIISNLSKAAGLASMIAAMRPDIDNIDEYVRNTTARAFSVVASALG 604
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+P+LL FLKAVCQSK+SWQARHTGI+I+QQIAIL+GC VLPHLR VEII+HGL D K
Sbjct 605 IPALLPFLKAVCQSKRSWQARHTGIKIVQQIAILIGCAVLPHLRSLVEIIEHGLSDENQK 664
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T+TAL+LAALAEAAAPYGIE+FD VL+PL
Sbjct 665 VRTITALSLAALAEAAAPYGIESFDSVLKPL 695
> sce:YMR288W HSH155; Hsh155p
Length=971
Score = 171 bits (433), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 86/151 (56%), Positives = 107/151 (70%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDED R G+EII+NL+ AGL T++ MRPDI++ DEYVRN T+RA AVV ALG
Sbjct 250 LLIDEDPMVRSTGQEIITNLSTVAGLKTILTVMRPDIENEDEYVRNVTSRAAAVVAKALG 309
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
V LL F+ A C S+KSW+ARHTGI+I+QQI ILLG GVL HL + I+ L D +
Sbjct 310 VNQLLPFINAACHSRKSWKARHTGIKIVQQIGILLGIGVLNHLTGLMSCIKDCLMDDHVP 369
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++ VTA L+ LAE + PYGIE F+ VL PL
Sbjct 370 VRIVTAHTLSTLAENSYPYGIEVFNVVLEPL 400
> ath:AT2G16950 TRN1; TRN1 (TRANSPORTIN 1); protein transporter
Length=891
Score = 36.2 bits (82), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 54/115 (46%), Gaps = 10/115 (8%)
Query 44 VRNTTARAFAVVVSALG---VPSLL-LFLKAVCQS-KKSWQARHTGIEIIQQIAILLGCG 98
+R +A A V+ + G +P+L+ L K + S ++W+ R + + IA G
Sbjct 368 LRKCSAAAIDVLSNVFGDEILPALMPLIQKNLSASGDEAWKQREAAVLALGAIAEGCMNG 427
Query 99 VLPHLRQFVEIIQHGLDDPVLKIKTVTALALAALA-----EAAAPYGIEAFDCVL 148
+ PHL + V + LDD I++++ L+ E+ P G E F+ VL
Sbjct 428 LYPHLSEIVAFLLPLLDDKFPLIRSISCWTLSRFGKYLIQESGNPKGYEQFEKVL 482
> sce:YGL195W GCN1, AAS103, NDR1; Gcn1p
Length=2672
Score = 32.0 bits (71), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 40/76 (52%), Gaps = 3/76 (3%)
Query 62 PSLLLFLKAVCQS--KKSWQARHTGIEIIQQIAILLGC-GVLPHLRQFVEIIQHGLDDPV 118
PSL L + + + +S + +I+ +AIL+ ++P+L+Q ++ ++ + DPV
Sbjct 1561 PSLALIIHIIHRGMHDRSANIKRKACKIVGNMAILVDTKDLIPYLQQLIDEVEIAMVDPV 1620
Query 119 LKIKTVTALALAALAE 134
+ A AL AL E
Sbjct 1621 PNTRATAARALGALVE 1636
> dre:100334568 phosphodiesterase 5A-like
Length=954
Score = 31.2 bits (69), Expect = 1.4, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 8 YARIEGREIISNLAKAAGLATMIATMRPDIDH 39
+A ++ +E+ SNL+ LA MIA++ D+DH
Sbjct 696 FAMLQTKELKSNLSSLEVLALMIASLSHDLDH 727
> ath:AT2G16960 importin beta-2 subunit family protein
Length=505
Score = 30.8 bits (68), Expect = 1.6, Method: Composition-based stats.
Identities = 25/106 (23%), Positives = 46/106 (43%), Gaps = 11/106 (10%)
Query 34 RPDIDHPDE----YVRNTTARAFAVVVSALGVPSLLLFLKAVCQSK------KSWQARHT 83
+PDID +R +A+ ++ + G +LL L + ++K ++W+ R
Sbjct 19 QPDIDQAQNDKEWNLRACSAKFIGILANVFG-DEILLTLMPLIEAKLSKFDDETWKEREA 77
Query 84 GIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLKIKTVTALAL 129
+ IA PHL + V I++ LDD ++ +T L
Sbjct 78 AVFAFGAIAEGCNSFFYPHLAEIVAILRRLLDDQSPLVRRITCWTL 123
> dre:793480 kcnk1, MGC165664, im:7150627, zgc:165664; potassium
channel, subfamily K, member 1; K04912 potassium channel
subfamily K member 1
Length=338
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 49 ARAFAVVVSALGVPSLLLFLKAVCQSKKSWQAR 81
+AF ++ S +G+P LLFL AV Q + R
Sbjct 130 GKAFCIIYSVVGIPFTLLFLTAVVQRIMEFSTR 162
> cpv:cgd6_270 hypothetical protein
Length=1330
Score = 30.0 bits (66), Expect = 3.2, Method: Composition-based stats.
Identities = 19/76 (25%), Positives = 34/76 (44%), Gaps = 1/76 (1%)
Query 33 MRPDIDHPDEYVRNTTARAFAVVVSALGVPSLLLFLKAVCQSKKSWQARHTGIEIIQQIA 92
+R D+ HP+EY+R +T R + + L+ + Q + S+ R + I I
Sbjct 105 LRNDLQHPNEYIRGSTLRLLCNLRFIKLIQPLIESILENLQHRHSY-VRRNAVMCIYSII 163
Query 93 ILLGCGVLPHLRQFVE 108
G ++P+ VE
Sbjct 164 KTFGVDIIPNAVDEVE 179
> hsa:1315 COPB1, COPB, DKFZp761K102, FLJ10341, FLJ46444, FLJ57957;
coatomer protein complex, subunit beta 1
Length=953
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 34 RPDIDHPDEYVRNTTARAFAVVVSALGVPSLLLFLKAVCQSKKSWQARH 82
R D+ HP+E++R +T R + A + L+ ++A + + S+ R+
Sbjct 106 RKDLQHPNEFIRGSTLRFLCKLKEAELLEPLMPAIRACLEHRHSYVRRN 154
> mmu:70349 Copb1, 2610019B04Rik; coatomer protein complex, subunit
beta 1
Length=953
Score = 29.6 bits (65), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 34 RPDIDHPDEYVRNTTARAFAVVVSALGVPSLLLFLKAVCQSKKSWQARH 82
R D+ HP+E++R +T R + A + L+ ++A + + S+ R+
Sbjct 106 RKDLQHPNEFIRGSTLRFLCKLKEAELLEPLMPAIRACLEHRHSYVRRN 154
> cel:Y48G9A.3 hypothetical protein
Length=2680
Score = 29.6 bits (65), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 8/95 (8%)
Query 46 NTTARAFAVVVSA-----LGVPSLLLFLKAVCQS--KKSWQARHTGIEIIQQI-AILLGC 97
N T+ A V++ + PSL L + V ++ + + R +II I ++
Sbjct 1605 NKTSAALQAVLNTKFIHYIDAPSLALMMPIVRRAFEDRLSETRRVAAQIISNIYSLTENK 1664
Query 98 GVLPHLRQFVEIIQHGLDDPVLKIKTVTALALAAL 132
+ P+L V +Q L DPV +I+ V+A AL A+
Sbjct 1665 DMEPYLAHMVPGLQRSLLDPVPEIRAVSARALGAV 1699
> xla:735158 hypothetical protein MGC114704
Length=953
Score = 29.6 bits (65), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 34 RPDIDHPDEYVRNTTARAFAVVVSALGVPSLLLFLKAVCQSKKSWQARH 82
R D+ HP+E++R +T R + A + L+ ++A + + S+ R+
Sbjct 106 RKDLQHPNEFIRGSTLRFLCKLKEAELLEPLMPAIRACLEHRHSYVRRN 154
> dre:100002604 phosphodiesterase 5A-like
Length=964
Score = 29.6 bits (65), Expect = 3.9, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 8 YARIEGREIISNLAKAAGLATMIATMRPDIDH 39
+A ++ +E+ SN + LA MIA++ D+DH
Sbjct 696 FAMLQTKELKSNFSSLEVLALMIASLSHDLDH 727
> hsa:79711 IPO4, FLJ23338, Imp4, MGC131665; importin 4
Length=1081
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 11/95 (11%)
Query 62 PSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQ-----FVEIIQHGLDD 116
P L+ L+ +S+ +Q R G+ ++ +L G H+RQ ++I+ GL+D
Sbjct 351 PQLMPMLEEALRSESPYQ-RKAGLLVL----AVLSDGAGDHIRQRLLPPLLQIVCKGLED 405
Query 117 PVLKIKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
P ++ AL +E P+ I ++ + PL
Sbjct 406 PSQVVRNAALFALGQFSENLQPH-ISSYSREVMPL 439
> hsa:3775 KCNK1, DPK, HOHO, K2P1, K2p1.1, KCNO1, TWIK-1, TWIK1;
potassium channel, subfamily K, member 1; K04912 potassium
channel subfamily K member 1
Length=336
Score = 29.6 bits (65), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 49 ARAFAVVVSALGVPSLLLFLKAVCQ 73
+AF ++ S +G+P LLFL AV Q
Sbjct 130 GKAFCIIYSVIGIPFTLLFLTAVVQ 154
> mmu:16525 Kcnk1, AI788889, TWIK-1; potassium channel, subfamily
K, member 1; K04912 potassium channel subfamily K member
1
Length=336
Score = 29.6 bits (65), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 49 ARAFAVVVSALGVPSLLLFLKAVCQ 73
+AF ++ S +G+P LLFL AV Q
Sbjct 130 GKAFCIIYSVIGIPFTLLFLTAVVQ 154
> ath:AT1G64790 binding
Length=2610
Score = 29.6 bits (65), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 23/121 (19%), Positives = 62/121 (51%), Gaps = 5/121 (4%)
Query 17 ISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAF-AVVVSALGVPSLLLFLKAVCQS- 74
+ ++ K +++++ T+ + P+EY R+ V+++ PSL L + V +
Sbjct 1508 VGSVIKNPEISSLVPTLLLALTDPNEYTRHALDTLLQTTFVNSVDAPSLALLVPIVHRGL 1567
Query 75 -KKSWQARHTGIEIIQQIAILLG--CGVLPHLRQFVEIIQHGLDDPVLKIKTVTALALAA 131
++S + + +I+ + L+ ++P++ + ++ L DP+ ++++V A A+ +
Sbjct 1568 RERSSETKKKASQIVGNMCSLVTEPKDMIPYIGLLLPEVKKVLVDPIPEVRSVAARAVGS 1627
Query 132 L 132
L
Sbjct 1628 L 1628
> dre:559366 lrrk2; leucine-rich repeat kinase 2 (EC:2.7.11.1);
K08844 leucine-rich repeat kinase 2 [EC:2.7.11.1]
Length=2533
Score = 29.3 bits (64), Expect = 4.4, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 5/46 (10%)
Query 81 RHTGI-EIIQQIAILLGCGVLPHLRQFVE---IIQHGLDDPVLKIK 122
RH+ + EI+Q+ + L G LPH F+ ++ H DDPVL++K
Sbjct 1536 RHSRLMEILQETQLQLEEGELPHAIHFLSEAGVLLH-FDDPVLQLK 1580
> dre:338138 copb1, MGC92482, id:ibd5034, wu:fc66c05, zgc:92482;
coatomer protein complex, subunit beta 1
Length=953
Score = 28.9 bits (63), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 12/49 (24%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 34 RPDIDHPDEYVRNTTARAFAVVVSALGVPSLLLFLKAVCQSKKSWQARH 82
R D+ HP+E++R +T R + + + L+ ++A + + S+ R+
Sbjct 106 RKDLQHPNEFIRGSTLRFLCKLKESELLEPLMPAIRACLERRHSYVRRN 154
> xla:444286 copb1, MGC80934; coatomer protein complex, subunit
beta 1
Length=960
Score = 28.9 bits (63), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 26/49 (53%), Gaps = 0/49 (0%)
Query 34 RPDIDHPDEYVRNTTARAFAVVVSALGVPSLLLFLKAVCQSKKSWQARH 82
R D+ HP+E++R +T R + A + L+ ++A + S+ R+
Sbjct 106 RKDLQHPNEFIRGSTLRFLCKLKEAELLEPLMPAIRACLDHRHSYVRRN 154
> mmu:75751 Ipo4, 8430408O15Rik, AA409693, Imp4a, MGC113723, RanBP4;
importin 4
Length=1082
Score = 28.5 bits (62), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 44/95 (46%), Gaps = 11/95 (11%)
Query 62 PSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQ-----FVEIIQHGLDD 116
P ++ L+ +S+ +Q R G ++ +L G H+RQ ++I+ GLDD
Sbjct 351 PHVMPMLEEALRSEDPYQ-RKAGFLVLA----VLSDGAGDHIRQRLLYPLLQIVCKGLDD 405
Query 117 PVLKIKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
P ++ AL +E P+ I ++ + PL
Sbjct 406 PSQIVRNAALFALGQFSENLQPH-ISSYSEEVMPL 439
Lambda K H
0.325 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3199347004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40