bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2510_orf1
Length=83
Score E
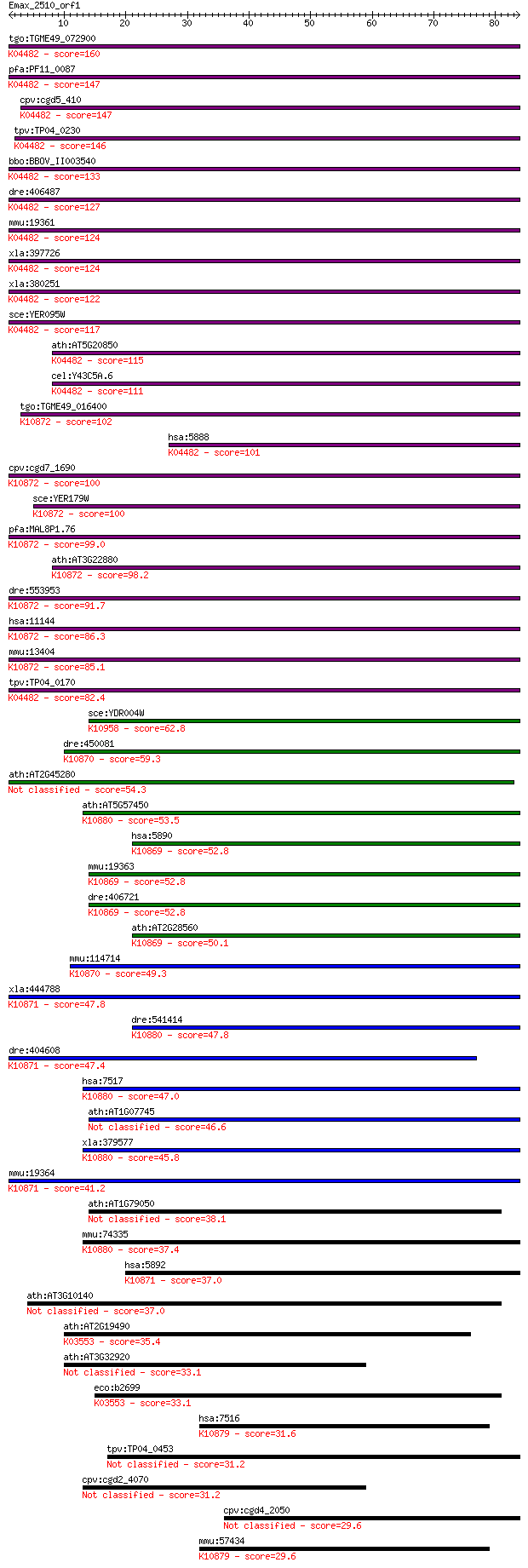
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8); K... 160 1e-39
pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein ... 147 6e-36
cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51 147 7e-36
tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair prot... 146 1e-35
bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair ... 133 2e-31
dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA h... 127 1e-29
mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cer... 124 7e-29
xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad... 124 8e-29
xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,... 122 2e-28
sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51 117 1e-26
ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-depende... 115 3e-26
cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-... 111 6e-25
tgo:TGME49_016400 meiotic recombination protein DMC1-like prot... 102 3e-22
hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA... 101 7e-22
cpv:cgd7_1690 meiotic recombination protein DMC1-like protein ... 100 8e-22
sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination pr... 100 2e-21
pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;... 99.0 3e-21
ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1); AT... 98.2 6e-21
dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor... 91.7 5e-19
hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473, d... 86.3 2e-17
mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage sup... 85.1 5e-17
tpv:TP04_0170 meiotic recombination protein DMC1; K04482 DNA r... 82.4 3e-16
sce:YDR004W RAD57; Protein that stimulates strand exchange by ... 62.8 3e-10
dre:450081 rad51c, zgc:101596; rad51 homolog C (S. cerevisiae)... 59.3 3e-09
ath:AT2G45280 ATRAD51C; ATP binding / damaged DNA binding / pr... 54.3 1e-07
ath:AT5G57450 XRCC3; XRCC3; ATP binding / damaged DNA binding ... 53.5 2e-07
hsa:5890 RAD51L1, MGC34245, R51H2, RAD51B, REC2, hREC2; RAD51-... 52.8 2e-07
mmu:19363 Rad51l1, AI553500, R51H2, Rad51b, mREC2; RAD51-like ... 52.8 3e-07
dre:406721 rad51l1, wu:fd07f04, zgc:56581; RAD51-like 1 (S. ce... 52.8 3e-07
ath:AT2G28560 RAD51B; RAD51B; recombinase; K10869 RAD51-like p... 50.1 2e-06
mmu:114714 Rad51c, R51H3, Rad51l2; RAD51 homolog c (S. cerevis... 49.3 3e-06
xla:444788 rad51l3, MGC82048; RAD51-like 3; K10871 RAD51-like ... 47.8 9e-06
dre:541414 xrcc3, im:7142103, si:dkey-11b8.1, zgc:101608; X-ra... 47.8 9e-06
dre:404608 MGC77165; zgc:77165; K10871 RAD51-like protein 3 47.4
hsa:7517 XRCC3, CMM6; X-ray repair complementing defective rep... 47.0 1e-05
ath:AT1G07745 RAD51D; RAD51D (ARABIDOPSIS HOMOLOG OF RAD51 D);... 46.6 2e-05
xla:379577 xrcc3, MGC69118; X-ray repair complementing defecti... 45.8 3e-05
mmu:19364 Rad51l3, DKFZp586D0122, R51H3, Rad51d, Trad-d2, Trad... 41.2 0.001
ath:AT1G79050 DNA repair protein recA 38.1 0.008
mmu:74335 Xrcc3, 4432412E01Rik, AI182522, AW537713; X-ray repa... 37.4 0.012
hsa:5892 RAD51L3, R51H3, RAD51D, TRAD; RAD51-like 3 (S. cerevi... 37.0 0.016
ath:AT3G10140 RECA3; RECA3 (recA homolog 3); ATP binding / DNA... 37.0 0.016
ath:AT2G19490 recA family protein; K03553 recombination protei... 35.4 0.046
ath:AT3G32920 ATP binding / DNA binding / DNA-dependent ATPase... 33.1 0.20
eco:b2699 recA, ECK2694, JW2669, lexB, recH, rnmB, srf, tif, u... 33.1 0.24
hsa:7516 XRCC2, DKFZp781P0919; X-ray repair complementing defe... 31.6 0.71
tpv:TP04_0453 hypothetical protein 31.2 0.79
cpv:cgd2_4070 hypothetical protein 31.2 0.89
cpv:cgd4_2050 hypothetical protein 29.6 2.7
mmu:57434 Xrcc2, 4921524O04Rik, 8030409M04Rik, RAD51, RecA; X-... 29.6 2.7
> tgo:TGME49_072900 DNA repair protein, putative (EC:3.6.3.8);
K04482 DNA repair protein RAD51
Length=354
Score = 160 bits (404), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 71/83 (85%), Positives = 80/83 (96%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
AQEYLEAR N+I+FTTGSVQLD+LLKGG+ETGN+TELFGEFR GKTQLCHT AV+CQLP+
Sbjct 102 AQEYLEARENLIRFTTGSVQLDSLLKGGIETGNLTELFGEFRTGKTQLCHTLAVTCQLPI 161
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
EQ+GGEGKCLWIDTEGTFRPER+
Sbjct 162 EQAGGEGKCLWIDTEGTFRPERI 184
> pfa:PF11_0087 rad51; Rad51 homolog; K04482 DNA repair protein
RAD51
Length=350
Score = 147 bits (372), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 66/83 (79%), Positives = 76/83 (91%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A +Y +AR N+IKFTTGS QLDALLKGG+ETG +TELFGEFR GK+QLCHT A++CQLP+
Sbjct 99 AIDYHDARQNLIKFTTGSKQLDALLKGGIETGGITELFGEFRTGKSQLCHTLAITCQLPI 158
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
EQSGGEGKCLWIDTEGTFRPER+
Sbjct 159 EQSGGEGKCLWIDTEGTFRPERI 181
> cpv:cgd5_410 Rad51 ; K04482 DNA repair protein RAD51
Length=347
Score = 147 bits (372), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 66/81 (81%), Positives = 74/81 (91%), Gaps = 0/81 (0%)
Query 3 EYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQ 62
EYLEAR N+IKFTTGS QLD LL+GG+ETG++TE+FGEFR GKTQLCHT AV+CQLPVE
Sbjct 97 EYLEARTNLIKFTTGSSQLDRLLQGGIETGSITEIFGEFRTGKTQLCHTLAVTCQLPVEH 156
Query 63 SGGEGKCLWIDTEGTFRPERV 83
GGEGKCLWIDTEGTFRPER+
Sbjct 157 KGGEGKCLWIDTEGTFRPERI 177
> tpv:TP04_0230 DNA repair protein Rad51; K04482 DNA repair protein
RAD51
Length=343
Score = 146 bits (369), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 64/82 (78%), Positives = 76/82 (92%), Gaps = 0/82 (0%)
Query 2 QEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVE 61
Q+YLEAR N+IKFTTGS QLD LL+GGVETG++TE+ GEF+ GK+QLCHT AV+CQLPVE
Sbjct 93 QDYLEARGNLIKFTTGSAQLDKLLQGGVETGSITEIIGEFKTGKSQLCHTLAVTCQLPVE 152
Query 62 QSGGEGKCLWIDTEGTFRPERV 83
QSGGEGKCLW+D+EGTFRPER+
Sbjct 153 QSGGEGKCLWVDSEGTFRPERI 174
> bbo:BBOV_II003540 18.m06297; Rad51 protein; K04482 DNA repair
protein RAD51
Length=346
Score = 133 bits (334), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 57/83 (68%), Positives = 72/83 (86%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A EYLE R N+IKFTTGS LDALL+GG+E+G++TE+ G+F GKTQLCHT A++ QLP+
Sbjct 92 AAEYLECRLNLIKFTTGSTALDALLQGGIESGSITEIIGDFSTGKTQLCHTLAITSQLPI 151
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
EQ+GGEGKCLWIDT+ +FRPER+
Sbjct 152 EQNGGEGKCLWIDTQNSFRPERL 174
> dre:406487 rad51, wu:fb38e12, zgc:77754; RAD51 homolog (RecA
homolog, E. coli) (S. cerevisiae); K04482 DNA repair protein
RAD51
Length=338
Score = 127 bits (318), Expect = 1e-29, Method: Composition-based stats.
Identities = 56/83 (67%), Positives = 72/83 (86%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A E+ + RA II+ +TGS +LD LL+GG+ETG++TE+FGEFR GKTQLCHT AV+CQLP+
Sbjct 88 ATEFHQRRAEIIQISTGSKELDKLLQGGIETGSITEMFGEFRTGKTQLCHTLAVTCQLPI 147
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
+Q GGEGK ++IDTEGTFRPER+
Sbjct 148 DQGGGEGKAMYIDTEGTFRPERL 170
> mmu:19361 Rad51, AV304093, Rad51a, Reca; RAD51 homolog (S. cerevisiae);
K04482 DNA repair protein RAD51
Length=339
Score = 124 bits (311), Expect = 7e-29, Method: Composition-based stats.
Identities = 54/83 (65%), Positives = 72/83 (86%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A E+ + R+ II+ TTGS +LD LL+GG+ETG++TE+FGEFR GKTQ+CHT AV+CQLP+
Sbjct 89 ATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQICHTLAVTCQLPI 148
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
++ GGEGK ++IDTEGTFRPER+
Sbjct 149 DRGGGEGKAMYIDTEGTFRPERL 171
> xla:397726 rad51-a, MGC84850, brcc5, rad51, rad51a, reca, xrad51;
RAD51 homolog (RecA homolog); K04482 DNA repair protein
RAD51
Length=336
Score = 124 bits (311), Expect = 8e-29, Method: Composition-based stats.
Identities = 55/83 (66%), Positives = 72/83 (86%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A E+ + R+ II+ +TGS +LD LL+GGVETG++TE+FGEFR GKTQLCHT AV+CQLP+
Sbjct 86 ATEFHQRRSEIIQISTGSKELDKLLQGGVETGSITEMFGEFRTGKTQLCHTLAVTCQLPI 145
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
++ GGEGK ++IDTEGTFRPER+
Sbjct 146 DRGGGEGKAMYIDTEGTFRPERL 168
> xla:380251 rad51-b, MGC130792, MGC52570, brcc5, rad51, rad51a,
reca, xrad51; RAD51 homolog (RecA homolog); K04482 DNA repair
protein RAD51
Length=336
Score = 122 bits (306), Expect = 2e-28, Method: Composition-based stats.
Identities = 54/83 (65%), Positives = 71/83 (85%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A E+ + R+ II+ TGS +LD LL+GG+ETG++TE+FGEFR GKTQLCHT AV+CQLP+
Sbjct 86 ATEFHQRRSEIIQIGTGSKELDKLLQGGIETGSITEMFGEFRTGKTQLCHTLAVTCQLPI 145
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
++ GGEGK ++IDTEGTFRPER+
Sbjct 146 DRGGGEGKAMYIDTEGTFRPERL 168
> sce:YER095W RAD51, MUT5; Rad51p; K04482 DNA repair protein RAD51
Length=400
Score = 117 bits (292), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 54/83 (65%), Positives = 67/83 (80%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A ++ R+ +I TTGS LD LL GGVETG++TELFGEFR GK+QLCHT AV+CQ+P+
Sbjct 147 AADFHMRRSELICLTTGSKNLDTLLGGGVETGSITELFGEFRTGKSQLCHTLAVTCQIPL 206
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
+ GGEGKCL+IDTEGTFRP R+
Sbjct 207 DIGGGEGKCLYIDTEGTFRPVRL 229
> ath:AT5G20850 ATRAD51; ATP binding / DNA binding / DNA-dependent
ATPase/ damaged DNA binding / nucleoside-triphosphatase/
nucleotide binding / protein binding / sequence-specific DNA
binding; K04482 DNA repair protein RAD51
Length=342
Score = 115 bits (288), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 49/76 (64%), Positives = 66/76 (86%), Gaps = 0/76 (0%)
Query 8 RANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEG 67
R II+ T+GS +LD +L+GG+ETG++TEL+GEFR+GKTQLCHT V+CQLP++Q GGEG
Sbjct 99 RQEIIQITSGSRELDKVLEGGIETGSITELYGEFRSGKTQLCHTLCVTCQLPMDQGGGEG 158
Query 68 KCLWIDTEGTFRPERV 83
K ++ID EGTFRP+R+
Sbjct 159 KAMYIDAEGTFRPQRL 174
> cel:Y43C5A.6 rad-51; RADiation sensitivity abnormal/yeast RAD-related
family member (rad-51); K04482 DNA repair protein
RAD51
Length=395
Score = 111 bits (277), Expect = 6e-25, Method: Composition-based stats.
Identities = 46/76 (60%), Positives = 62/76 (81%), Gaps = 0/76 (0%)
Query 8 RANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEG 67
R+ +++ TGS LD LL GG+ETG++TE++GE+R GKTQLCH+ AV CQLP++ GGEG
Sbjct 150 RSQLVQIRTGSASLDRLLGGGIETGSITEVYGEYRTGKTQLCHSLAVLCQLPIDMGGGEG 209
Query 68 KCLWIDTEGTFRPERV 83
KC++IDT TFRPER+
Sbjct 210 KCMYIDTNATFRPERI 225
> tgo:TGME49_016400 meiotic recombination protein DMC1-like protein,
putative (EC:2.7.11.1); K10872 meiotic recombination
protein DMC1
Length=349
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 50/81 (61%), Positives = 60/81 (74%), Gaps = 0/81 (0%)
Query 3 EYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQ 62
E ++ R +IK TTGS QLD LL GG ET ++TELFGE R GKTQLCHT V+ QLP +
Sbjct 101 ELVQKRGRVIKITTGSDQLDQLLGGGFETMSITELFGENRCGKTQLCHTVCVTAQLPRDM 160
Query 63 SGGEGKCLWIDTEGTFRPERV 83
GG GK +IDTEGTFRPE++
Sbjct 161 KGGCGKVCYIDTEGTFRPEKI 181
> hsa:5888 RAD51, BRCC5, HRAD51, HsRad51, HsT16930, RAD51A, RECA;
RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae); K04482
DNA repair protein RAD51
Length=340
Score = 101 bits (251), Expect = 7e-22, Method: Composition-based stats.
Identities = 41/57 (71%), Positives = 53/57 (92%), Gaps = 0/57 (0%)
Query 27 GGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTFRPERV 83
GG+ETG++TE+FGEFR GKTQ+CHT AV+CQLP+++ GGEGK ++IDTEGTFRPER+
Sbjct 116 GGIETGSITEMFGEFRTGKTQICHTLAVTCQLPIDRGGGEGKAMYIDTEGTFRPERL 172
> cpv:cgd7_1690 meiotic recombination protein DMC1-like protein
; K10872 meiotic recombination protein DMC1
Length=342
Score = 100 bits (250), Expect = 8e-22, Method: Composition-based stats.
Identities = 47/83 (56%), Positives = 60/83 (72%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
E + R NI++ TTGS Q D +L GG E+ +TE+FGE R GKTQ+CHT V+ QLP+
Sbjct 92 GSEVMSRRQNILRITTGSEQFDKMLMGGFESMCITEIFGENRCGKTQICHTLCVAAQLPL 151
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
E +GG GK +IDTEGTFRPER+
Sbjct 152 EMNGGNGKVCFIDTEGTFRPERI 174
> sce:YER179W DMC1, ISC2; Dmc1p; K10872 meiotic recombination
protein DMC1
Length=334
Score = 100 bits (248), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 46/79 (58%), Positives = 59/79 (74%), Gaps = 0/79 (0%)
Query 5 LEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSG 64
L+ R + +TGS QLD++L GG+ T ++TE+FGEFR GKTQ+ HT V+ QLP E G
Sbjct 87 LDIRQRVYSLSTGSKQLDSILGGGIMTMSITEVFGEFRCGKTQMSHTLCVTTQLPREMGG 146
Query 65 GEGKCLWIDTEGTFRPERV 83
GEGK +IDTEGTFRPER+
Sbjct 147 GEGKVAYIDTEGTFRPERI 165
> pfa:MAL8P1.76 meiotic recombination protein dmc1-like protein;
K10872 meiotic recombination protein DMC1
Length=347
Score = 99.0 bits (245), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 47/83 (56%), Positives = 60/83 (72%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A E ++ R+ ++K TTGS D L GG+E+ +TELFGE R GKTQ+CHT AV+ QLP
Sbjct 97 ANELVQKRSKVLKITTGSTVFDQTLGGGIESMCITELFGENRCGKTQVCHTLAVTAQLPK 156
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
+GG GK +IDTEGTFRPE+V
Sbjct 157 SLNGGNGKVCYIDTEGTFRPEKV 179
> ath:AT3G22880 DMC1; DMC1 (DISRUPTION OF MEIOTIC CONTROL 1);
ATP binding / DNA binding / DNA-dependent ATPase/ damaged DNA
binding / nucleoside-triphosphatase/ nucleotide binding /
protein binding; K10872 meiotic recombination protein DMC1
Length=344
Score = 98.2 bits (243), Expect = 6e-21, Method: Composition-based stats.
Identities = 45/76 (59%), Positives = 55/76 (72%), Gaps = 0/76 (0%)
Query 8 RANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEG 67
R +++K TTG LD LL GG+ET +TE FGEFR+GKTQL HT V+ QLP GG G
Sbjct 102 RKSVVKITTGCQALDDLLGGGIETSAITEAFGEFRSGKTQLAHTLCVTTQLPTNMKGGNG 161
Query 68 KCLWIDTEGTFRPERV 83
K +IDTEGTFRP+R+
Sbjct 162 KVAYIDTEGTFRPDRI 177
> dre:553953 dmc1, MGC136628, zgc:136628; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=342
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 46/83 (55%), Positives = 55/83 (66%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A EY R + TTGS++ D LL GGVE+ +TE FGEFR GKTQL HT V+ QLP
Sbjct 90 ASEYCIKRKQVFHITTGSLEFDKLLGGGVESMAITEAFGEFRTGKTQLSHTLCVTAQLPG 149
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
E GK ++IDTE TFRPER+
Sbjct 150 EYGYTGGKVIFIDTENTFRPERL 172
> hsa:11144 DMC1, DMC1H, HsLim15, LIM15, MGC150472, MGC150473,
dJ199H16.1; DMC1 dosage suppressor of mck1 homolog, meiosis-specific
homologous recombination (yeast); K10872 meiotic recombination
protein DMC1
Length=340
Score = 86.3 bits (212), Expect = 2e-17, Method: Composition-based stats.
Identities = 44/83 (53%), Positives = 54/83 (65%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A EY E R + TTGS + D LL GG+E+ +TE FGEFR GKTQL HT V+ QLP
Sbjct 88 AFEYSEKRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTAQLPG 147
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
GK ++IDTE TFRP+R+
Sbjct 148 AGGYPGGKIIFIDTENTFRPDRL 170
> mmu:13404 Dmc1, Dmc1h, MGC151144, Mei11, sgdp; DMC1 dosage suppressor
of mck1 homolog, meiosis-specific homologous recombination
(yeast); K10872 meiotic recombination protein DMC1
Length=340
Score = 85.1 bits (209), Expect = 5e-17, Method: Composition-based stats.
Identities = 43/83 (51%), Positives = 54/83 (65%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A +Y E R + TTGS + D LL GG+E+ +TE FGEFR GKTQL HT V+ QLP
Sbjct 88 AFQYSERRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTAQLPG 147
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
GK ++IDTE TFRP+R+
Sbjct 148 TGGYSGGKIIFIDTENTFRPDRL 170
> tpv:TP04_0170 meiotic recombination protein DMC1; K04482 DNA
repair protein RAD51
Length=346
Score = 82.4 bits (202), Expect = 3e-16, Method: Composition-based stats.
Identities = 43/85 (50%), Positives = 57/85 (67%), Gaps = 6/85 (7%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQL-- 58
A E + R +I+K TGS L+ LL GG+ET ++TELFGE R GKTQ+CHT +V+ Q+
Sbjct 98 ASELYKIRKSILKINTGSEMLNRLLNGGIETMSITELFGENRTGKTQICHTISVTSQIIN 157
Query 59 PVEQSGGEGKCLWIDTEGTFRPERV 83
P E K +IDTE TFRPE++
Sbjct 158 PTEPF----KVCYIDTENTFRPEKI 178
> sce:YDR004W RAD57; Protein that stimulates strand exchange by
stabilizing the binding of Rad51p to single-stranded DNA;
involved in the recombinational repair of double-strand breaks
in DNA during vegetative growth and meiosis; forms heterodimer
with Rad55p; K10958 DNA repair protein RAD57
Length=460
Score = 62.8 bits (151), Expect = 3e-10, Method: Composition-based stats.
Identities = 32/70 (45%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWID 73
FTT V +D LL GG+ T +TE+FGE GK+QL A+S QL G GKC++I
Sbjct 100 FTTADVAMDELLGGGIFTHGITEIFGESSTGKSQLLMQLALSVQLSEPAGGLGGKCVYIT 159
Query 74 TEGTFRPERV 83
TEG +R+
Sbjct 160 TEGDLPTQRL 169
> dre:450081 rad51c, zgc:101596; rad51 homolog C (S. cerevisiae);
K10870 RAD51-like protein 2
Length=362
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 35/74 (47%), Positives = 44/74 (59%), Gaps = 3/74 (4%)
Query 10 NIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKC 69
+I+ F +G LD + GGV G TE+ G GKTQLC AV Q+PV G GK
Sbjct 85 SIVTFCSG---LDDAIGGGVPVGKTTEICGAPGVGKTQLCMQLAVDVQIPVFFGGLGGKA 141
Query 70 LWIDTEGTFRPERV 83
L+IDTEG+F +RV
Sbjct 142 LYIDTEGSFLVQRV 155
> ath:AT2G45280 ATRAD51C; ATP binding / damaged DNA binding /
protein binding / recombinase/ single-stranded DNA binding
Length=363
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 31/82 (37%), Positives = 47/82 (57%), Gaps = 0/82 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A + L ++ + TT LD +L GG+ ++TE+ G GKTQ+ +V+ Q+P
Sbjct 92 AWDMLHEEESLPRITTSCSDLDNILGGGISCRDVTEIGGVPGIGKTQIGIQLSVNVQIPR 151
Query 61 EQSGGEGKCLWIDTEGTFRPER 82
E G GK ++IDTEG+F ER
Sbjct 152 ECGGLGGKAIYIDTEGSFMVER 173
> ath:AT5G57450 XRCC3; XRCC3; ATP binding / damaged DNA binding
/ protein binding / single-stranded DNA binding; K10880 DNA-repair
protein XRCC3
Length=304
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/71 (38%), Positives = 39/71 (54%), Gaps = 0/71 (0%)
Query 13 KFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWI 72
K TTG LD L+GG+ ++TE+ E GKTQLC ++ QLP+ G G L++
Sbjct 20 KLTTGCEILDGCLRGGISCDSLTEIVAESGCGKTQLCLQLSLCTQLPISHGGLNGSSLYL 79
Query 73 DTEGTFRPERV 83
+E F R+
Sbjct 80 HSEFPFPFRRL 90
> hsa:5890 RAD51L1, MGC34245, R51H2, RAD51B, REC2, hREC2; RAD51-like
1 (S. cerevisiae); K10869 RAD51-like protein 1
Length=350
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/63 (42%), Positives = 35/63 (55%), Gaps = 0/63 (0%)
Query 21 LDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTFRP 80
LD L GGV G++TE+ G GKTQ C ++ LP G EG ++IDTE F
Sbjct 90 LDEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYIDTESAFSA 149
Query 81 ERV 83
ER+
Sbjct 150 ERL 152
> mmu:19363 Rad51l1, AI553500, R51H2, Rad51b, mREC2; RAD51-like
1 (S. cerevisiae); K10869 RAD51-like protein 1
Length=350
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 37/70 (52%), Gaps = 0/70 (0%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWID 73
+T LD L GGV G++TE+ G GKTQ C +V LP G EG ++ID
Sbjct 83 LSTTLCALDEALHGGVPCGSLTEITGPPGCGKTQFCIMMSVLATLPTSLGGLEGAVVYID 142
Query 74 TEGTFRPERV 83
TE F ER+
Sbjct 143 TESAFTAERL 152
> dre:406721 rad51l1, wu:fd07f04, zgc:56581; RAD51-like 1 (S.
cerevisiae); K10869 RAD51-like protein 1
Length=373
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/70 (41%), Positives = 38/70 (54%), Gaps = 0/70 (0%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWID 73
F+T LD LL GG+ G +TE+ G GKTQLC +V LP G + ++ID
Sbjct 79 FSTSLPALDRLLHGGLPRGALTEVTGPSGCGKTQLCMMLSVLATLPKSLGGLDSGVIYID 138
Query 74 TEGTFRPERV 83
TE F ER+
Sbjct 139 TESAFSAERL 148
> ath:AT2G28560 RAD51B; RAD51B; recombinase; K10869 RAD51-like
protein 1
Length=370
Score = 50.1 bits (118), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 21 LDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTFRP 80
LD L GG+ G +TEL G GK+Q C A+S PV G +G+ ++ID E F
Sbjct 91 LDDTLCGGIPFGVLTELVGPPGIGKSQFCMKLALSASFPVAYGGLDGRVIYIDVESKFSS 150
Query 81 ERV 83
RV
Sbjct 151 RRV 153
> mmu:114714 Rad51c, R51H3, Rad51l2; RAD51 homolog c (S. cerevisiae);
K10870 RAD51-like protein 2
Length=366
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 31/73 (42%), Positives = 41/73 (56%), Gaps = 3/73 (4%)
Query 11 IIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCL 70
II F + LD +L GG+ TE+ G GKTQLC AV Q+P G G+ +
Sbjct 91 IITFCSA---LDNILGGGIPLMKTTEVCGVPGVGKTQLCMQLAVDVQIPECFGGVAGEAV 147
Query 71 WIDTEGTFRPERV 83
+IDTEG+F +RV
Sbjct 148 FIDTEGSFMVDRV 160
> xla:444788 rad51l3, MGC82048; RAD51-like 3; K10871 RAD51-like
protein 3
Length=324
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 27/83 (32%), Positives = 45/83 (54%), Gaps = 5/83 (6%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A Y E +++ T + +LD LL G+ TG +TE+ G +GKTQ+C + AV+ +
Sbjct 69 ADVYEELKSSTAILPTANRKLDILLDSGLYTGEVTEIAGAAGSGKTQMCQSIAVNVAYSL 128
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
+Q+ L++DT G R+
Sbjct 129 KQT-----VLYVDTTGGLTASRL 146
> dre:541414 xrcc3, im:7142103, si:dkey-11b8.1, zgc:101608; X-ray
repair complementing defective repair in Chinese hamster
cells 3; K10880 DNA-repair protein XRCC3
Length=352
Score = 47.8 bits (112), Expect = 9e-06, Method: Composition-based stats.
Identities = 25/63 (39%), Positives = 35/63 (55%), Gaps = 0/63 (0%)
Query 21 LDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTFRP 80
LD L++GG+ +TEL GE GKTQ C +S Q P E G ++I TE +F
Sbjct 89 LDGLMRGGLPLRGITELAGESAAGKTQFCLQLCLSVQYPQEHGGLNSGAVYICTEDSFPI 148
Query 81 ERV 83
+R+
Sbjct 149 KRL 151
> dre:404608 MGC77165; zgc:77165; K10871 RAD51-like protein 3
Length=327
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/76 (38%), Positives = 44/76 (57%), Gaps = 5/76 (6%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A Y E ++ +TGS LD LL G+ TG +TEL G +GKTQ+C + AV+ +
Sbjct 69 ADLYEELLSSTAILSTGSPSLDKLLDSGLYTGEITELTGSPGSGKTQVCFSVAVNISHQL 128
Query 61 EQSGGEGKCLWIDTEG 76
+Q+ ++IDT+G
Sbjct 129 KQT-----VVYIDTKG 139
> hsa:7517 XRCC3, CMM6; X-ray repair complementing defective repair
in Chinese hamster cells 3; K10880 DNA-repair protein
XRCC3
Length=346
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 38/71 (53%), Gaps = 0/71 (0%)
Query 13 KFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWI 72
+ + G LDALL+GG+ +TEL G GKTQL ++ Q P + G E ++I
Sbjct 81 RLSLGCPVLDALLRGGLPLDGITELAGRSSAGKTQLALQLCLAVQFPRQHGGLEAGAVYI 140
Query 73 DTEGTFRPERV 83
TE F +R+
Sbjct 141 CTEDAFPHKRL 151
> ath:AT1G07745 RAD51D; RAD51D (ARABIDOPSIS HOMOLOG OF RAD51 D);
ATP binding / DNA binding / DNA-dependent ATPase
Length=304
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 25/70 (35%), Positives = 36/70 (51%), Gaps = 5/70 (7%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWID 73
+TG + D+LL+GG G +TEL G +GKTQ C A S G+ L++D
Sbjct 71 LSTGDKETDSLLQGGFREGQLTELVGPSSSGKTQFCMQAAASV-----AENHLGRVLYLD 125
Query 74 TEGTFRPERV 83
T +F R+
Sbjct 126 TGNSFSARRI 135
> xla:379577 xrcc3, MGC69118; X-ray repair complementing defective
repair in Chinese hamster cells 3; K10880 DNA-repair protein
XRCC3
Length=350
Score = 45.8 bits (107), Expect = 3e-05, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 37/71 (52%), Gaps = 0/71 (0%)
Query 13 KFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWI 72
K + G LD L+GG+ +TE+ GE GKTQ+ +S Q PVE G ++I
Sbjct 81 KLSLGCKVLDNFLRGGIPLVGITEIAGESSAGKTQIGLQLCLSVQYPVEYGGLASGAVYI 140
Query 73 DTEGTFRPERV 83
TE F +R+
Sbjct 141 CTEDAFPSKRL 151
> mmu:19364 Rad51l3, DKFZp586D0122, R51H3, Rad51d, Trad-d2, Trad-d3,
Trad-d4, Trad-d6, Trad-d7; RAD51-like 3 (S. cerevisiae);
K10871 RAD51-like protein 3
Length=329
Score = 41.2 bits (95), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 42/83 (50%), Gaps = 5/83 (6%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A Y E + + +TG LD LL G+ TG +TE+ G +GKTQ+C A + +
Sbjct 69 ADLYEELKTSTAILSTGIGSLDKLLDAGLYTGEVTEIVGGPGSGKTQVCLCVAANVAHSL 128
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
+Q+ L++D+ G R+
Sbjct 129 QQN-----VLYVDSNGGMTASRL 146
> ath:AT1G79050 DNA repair protein recA
Length=343
Score = 38.1 bits (87), Expect = 0.008, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 6/67 (8%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWID 73
F++G + LD L GG+ G + E++G +GKT L HA++ V++ GG + +D
Sbjct 117 FSSGILTLDLALGGGLPKGRVVEIYGPESSGKTTLA-LHAIA---EVQKLGGNA--MLVD 170
Query 74 TEGTFRP 80
E F P
Sbjct 171 AEHAFDP 177
> mmu:74335 Xrcc3, 4432412E01Rik, AI182522, AW537713; X-ray repair
complementing defective repair in Chinese hamster cells
3; K10880 DNA-repair protein XRCC3
Length=349
Score = 37.4 bits (85), Expect = 0.012, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 0/71 (0%)
Query 13 KFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWI 72
+ + G LD L GG+ +T L G GKTQL ++ Q P + G E ++I
Sbjct 81 RLSLGCPVLDQFLGGGLPLEGITGLAGCSSAGKTQLALQLCLAVQFPRQYGGLEAGAVYI 140
Query 73 DTEGTFRPERV 83
TE F +R+
Sbjct 141 CTEDAFPSKRL 151
> hsa:5892 RAD51L3, R51H3, RAD51D, TRAD; RAD51-like 3 (S. cerevisiae);
K10871 RAD51-like protein 3
Length=348
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 35/64 (54%), Gaps = 5/64 (7%)
Query 20 QLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTFR 79
+LD LL G+ TG +TE+ G +GKTQ+C A + ++Q+ L++D+ G
Sbjct 108 RLDKLLDAGLYTGEVTEIVGGPGSGKTQVCLCMAANVAHGLQQN-----VLYVDSNGGLT 162
Query 80 PERV 83
R+
Sbjct 163 ASRL 166
> ath:AT3G10140 RECA3; RECA3 (recA homolog 3); ATP binding / DNA
binding / DNA-dependent ATPase/ nucleoside-triphosphatase/
nucleotide binding
Length=389
Score = 37.0 bits (84), Expect = 0.016, Method: Composition-based stats.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 9/78 (11%)
Query 4 YLEARANIIKFTTGSVQLD-ALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQ 62
Y + R ++I +TGS+ LD AL GG+ G M E++G+ +GKT L + + E
Sbjct 89 YRKRRVSVI--STGSLNLDLALGVGGLPKGRMVEVYGKEASGKTTL------ALHIIKEA 140
Query 63 SGGEGKCLWIDTEGTFRP 80
G C ++D E P
Sbjct 141 QKLGGYCAYLDAENAMDP 158
> ath:AT2G19490 recA family protein; K03553 recombination protein
RecA
Length=430
Score = 35.4 bits (80), Expect = 0.046, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 36/67 (53%), Gaps = 7/67 (10%)
Query 10 NIIKFTTGSVQLD-ALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGK 68
N+ F+TGS LD AL GG+ G + E++G +GKT L H ++ E G
Sbjct 89 NVPVFSTGSFALDVALGVGGLPKGRVVEIYGPEASGKTTLA-LHVIA-----EAQKQGGT 142
Query 69 CLWIDTE 75
C+++D E
Sbjct 143 CVFVDAE 149
> ath:AT3G32920 ATP binding / DNA binding / DNA-dependent ATPase/
nucleoside-triphosphatase/ nucleotide binding
Length=226
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Query 10 NIIKFTTGSVQLD-ALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQL 58
N+ F+TGS LD AL GG+ G + E++G +GKT L H +S L
Sbjct 11 NVPVFSTGSFALDVALGVGGLPKGRLVEIYGPEASGKTALA-LHMLSMLL 59
> eco:b2699 recA, ECK2694, JW2669, lexB, recH, rnmB, srf, tif,
umuB, umuR, zab; DNA strand exchange and recombination protein
with protease and nuclease activity; K03553 recombination
protein RecA
Length=353
Score = 33.1 bits (74), Expect = 0.24, Method: Composition-based stats.
Identities = 25/68 (36%), Positives = 34/68 (50%), Gaps = 9/68 (13%)
Query 15 TTGSVQLD-ALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGK-CLWI 72
+TGS+ LD AL GG+ G + E++G +GKT L + Q EGK C +I
Sbjct 42 STGSLSLDIALGAGGLPMGRIVEIYGPESSGKTTLTLQVIAAAQR-------EGKTCAFI 94
Query 73 DTEGTFRP 80
D E P
Sbjct 95 DAEHALDP 102
> hsa:7516 XRCC2, DKFZp781P0919; X-ray repair complementing defective
repair in Chinese hamster cells 2; K10879 DNA-repair
protein XRCC2
Length=280
Score = 31.6 bits (70), Expect = 0.71, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 32 GNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTF 78
G++ E G GKT++ + C LP + G E + L+IDT+ F
Sbjct 41 GDILEFHGPEGTGKTEMLYHLTARCILPKSEGGLEVEVLFIDTDYHF 87
> tpv:TP04_0453 hypothetical protein
Length=286
Score = 31.2 bits (69), Expect = 0.79, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 32/67 (47%), Gaps = 0/67 (0%)
Query 17 GSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEG 76
G ++D L GG+ G + E++G +GKTQ + + + L+I T G
Sbjct 31 GVKEIDQALNGGLLLGKVCEIYGPSGSGKTQFALSLTSEVLINNLIHSKDYVVLYIYTNG 90
Query 77 TFRPERV 83
TF ER+
Sbjct 91 TFPIERL 97
> cpv:cgd2_4070 hypothetical protein
Length=304
Score = 31.2 bits (69), Expect = 0.89, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 23/46 (50%), Gaps = 0/46 (0%)
Query 13 KFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQL 58
+ +TGS +D GG+ + E+ GE GKTQ C T S L
Sbjct 37 RLSTGSNVVDKAFNGGIPKRILFEITGEAGTGKTQWCLTLITSVLL 82
> cpv:cgd4_2050 hypothetical protein
Length=133
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 36 ELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWI-DTEGTFRPERV 83
EL G +GKT LC A++ Q+P G ++I D+EG F R+
Sbjct 13 ELCGVPGSGKTLLCKILALNIQIPKSIGGPGLNAIYIGDSEGGFSDNRL 61
> mmu:57434 Xrcc2, 4921524O04Rik, 8030409M04Rik, RAD51, RecA;
X-ray repair complementing defective repair in Chinese hamster
cells 2; K10879 DNA-repair protein XRCC2
Length=278
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 15/47 (31%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 32 GNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTF 78
G++ E G GKT++ + C LP + G + + L+IDT+ F
Sbjct 41 GDIFEFHGPEGTGKTEMLYHLTARCILPKSEGGLQIEVLFIDTDYHF 87
Lambda K H
0.317 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044675024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40