bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2580_orf1
Length=87
Score E
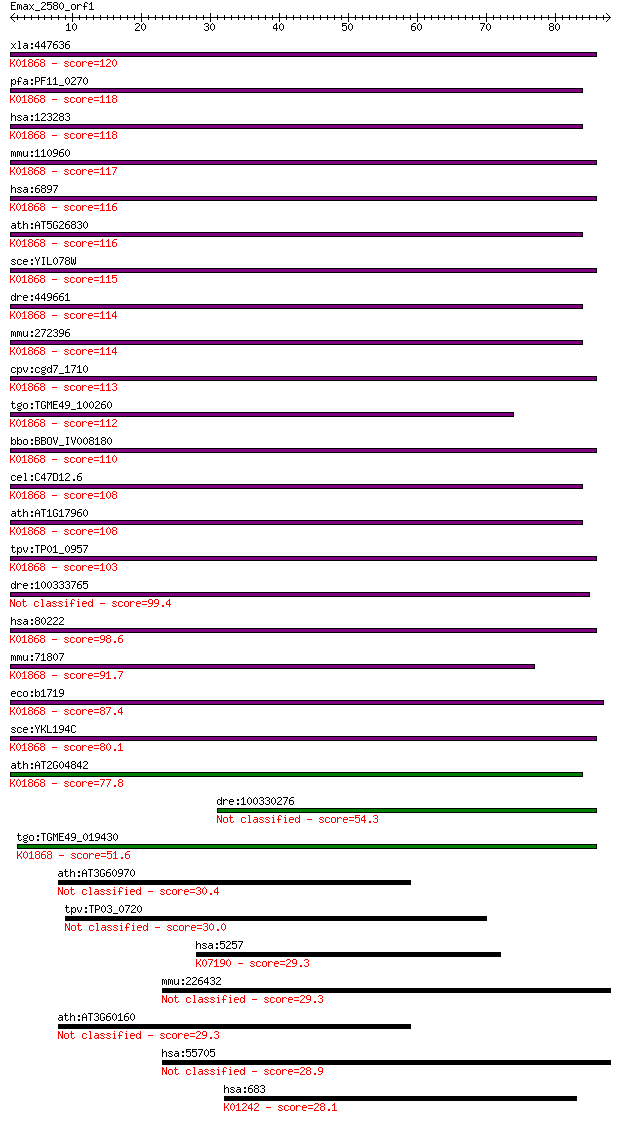
Sequences producing significant alignments: (Bits) Value
xla:447636 tars, MGC86352; threonyl-tRNA synthetase (EC:6.1.1.... 120 1e-27
pfa:PF11_0270 threonine --tRNA ligase, putative; K01868 threon... 118 4e-27
hsa:123283 TARSL2, FLJ25005; threonyl-tRNA synthetase-like 2 (... 118 4e-27
mmu:110960 Tars, D15Wsu59e, ThrRS; threonyl-tRNA synthetase (E... 117 7e-27
hsa:6897 TARS, MGC9344, ThrRS; threonyl-tRNA synthetase (EC:6.... 116 2e-26
ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligas... 116 2e-26
sce:YIL078W THS1; Ths1p (EC:6.1.1.3); K01868 threonyl-tRNA syn... 115 3e-26
dre:449661 tars, ThrRS, wu:fb07c05, wu:fb39c12, zgc:92586; thr... 114 8e-26
mmu:272396 Tarsl2, A530046H20Rik, MGC31414; threonyl-tRNA synt... 114 9e-26
cpv:cgd7_1710 threonyl-tRNA synthetase (RNA binding domain TGS... 113 2e-25
tgo:TGME49_100260 threonyl-tRNA synthetase, putative (EC:6.1.1... 112 2e-25
bbo:BBOV_IV008180 23.m05860; threonyl-tRNA synthetase (EC:6.1.... 110 1e-24
cel:C47D12.6 trs-1; Threonyl tRNA Synthetase family member (tr... 108 3e-24
ath:AT1G17960 threonyl-tRNA synthetase, putative / threonine--... 108 5e-24
tpv:TP01_0957 threonyl-tRNA synthetase; K01868 threonyl-tRNA s... 103 1e-22
dre:100333765 Threonyl-tRNA synthetase, cytoplasmic-like 99.4 2e-21
hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase 2,... 98.6 5e-21
mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRN... 91.7 5e-19
eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:... 87.4 1e-17
sce:YKL194C MST1; Mitochondrial threonyl-tRNA synthetase (EC:6... 80.1 2e-15
ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding / a... 77.8 8e-15
dre:100330276 threonyl-tRNA synthetase-like 54.3 9e-08
tgo:TGME49_019430 threonyl-tRNA synthetase, putative (EC:6.1.1... 51.6 5e-07
ath:AT3G60970 ATMRP15; ATPase, coupled to transmembrane moveme... 30.4 1.5
tpv:TP03_0720 hypothetical protein 30.0 1.7
hsa:5257 PHKB, DKFZp781E15103, FLJ41698; phosphorylase kinase,... 29.3 3.2
mmu:226432 Ipo9, 0710008K06Rik, AI845704, C78347, Imp9, Ranbp9... 29.3 3.4
ath:AT3G60160 ATMRP9; ATMRP9; ATPase, coupled to transmembrane... 29.3 3.5
hsa:55705 IPO9, DKFZp761M1547, FLJ10402, Imp9; importin 9 28.9
hsa:683 BST1, CD157; bone marrow stromal cell antigen 1 (EC:3.... 28.1 6.8
> xla:447636 tars, MGC86352; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=721
Score = 120 bits (300), Expect = 1e-27, Method: Composition-based stats.
Identities = 56/85 (65%), Positives = 67/85 (78%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFC +EQ+EEE S L+FL VY FGF+F L LST
Sbjct 439 HRNELSGALTGLTRVRRFQQDDAHIFCAMEQIEEEIKSCLDFLRSVYNVFGFTFKLNLST 498
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP K +G IE WNQAE L+N+L +
Sbjct 499 RPEKFLGDIEVWNQAEKQLENSLNE 523
> pfa:PF11_0270 threonine --tRNA ligase, putative; K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=1013
Score = 118 bits (296), Expect = 4e-27, Method: Composition-based stats.
Identities = 54/83 (65%), Positives = 65/83 (78%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SGSLSGLTRVRRFQQDD+HIFC++E +++E + L FLFYVY FGF + L LST
Sbjct 650 HRNEISGSLSGLTRVRRFQQDDSHIFCSMEHIKQEVLNTLNFLFYVYNLFGFKYELFLST 709
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP K IG I TWN AE L++AL
Sbjct 710 RPKKFIGQISTWNLAEQHLKDAL 732
> hsa:123283 TARSL2, FLJ25005; threonyl-tRNA synthetase-like 2
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=802
Score = 118 bits (296), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 57/83 (68%), Positives = 65/83 (78%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+LSGLTRVRRFQQDDAHIFCT+EQ+EEE L+FL VY FGFSF L LST
Sbjct 520 HRNELSGTLSGLTRVRRFQQDDAHIFCTVEQIEEEIKGCLQFLQSVYSTFGFSFQLNLST 579
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP +G IE WN+AE LQN+L
Sbjct 580 RPENFLGEIEMWNEAEKQLQNSL 602
> mmu:110960 Tars, D15Wsu59e, ThrRS; threonyl-tRNA synthetase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=722
Score = 117 bits (294), Expect = 7e-27, Method: Composition-based stats.
Identities = 55/85 (64%), Positives = 66/85 (77%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFC +EQ+E+E L+FL VY FGFSF L LST
Sbjct 440 HRNELSGALTGLTRVRRFQQDDAHIFCAMEQIEDEIKGCLDFLRTVYSVFGFSFKLNLST 499
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP K +G IE WNQAE L+N+L +
Sbjct 500 RPEKFLGDIEIWNQAEKQLENSLNE 524
> hsa:6897 TARS, MGC9344, ThrRS; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=723
Score = 116 bits (290), Expect = 2e-26, Method: Composition-based stats.
Identities = 54/85 (63%), Positives = 66/85 (77%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFC +EQ+E+E L+FL VY FGFSF L LST
Sbjct 441 HRNELSGALTGLTRVRRFQQDDAHIFCAMEQIEDEIKGCLDFLRTVYSVFGFSFKLNLST 500
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP K +G IE W+QAE L+N+L +
Sbjct 501 RPEKFLGDIEVWDQAEKQLENSLNE 525
> ath:AT5G26830 threonyl-tRNA synthetase / threonine--tRNA ligase
(THRRS) (EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=709
Score = 116 bits (290), Expect = 2e-26, Method: Composition-based stats.
Identities = 54/83 (65%), Positives = 67/83 (80%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+EASG+LSGLTRVRRFQQDDAHIFCT EQV+ E LEF+ YVY+ FGF++ L LST
Sbjct 435 HRNEASGALSGLTRVRRFQQDDAHIFCTTEQVKGEVQGVLEFIDYVYKVFGFTYELKLST 494
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP K +G +ETW++AE L+ A+
Sbjct 495 RPEKYLGDLETWDKAEADLKEAI 517
> sce:YIL078W THS1; Ths1p (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=734
Score = 115 bits (288), Expect = 3e-26, Method: Composition-based stats.
Identities = 53/85 (62%), Positives = 66/85 (77%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+LSGLTRVRRFQQDDAHIFCT +Q+E E + FL Y+Y FGF F + LST
Sbjct 448 HRNEFSGALSGLTRVRRFQQDDAHIFCTHDQIESEIENIFNFLQYIYGVFGFEFKMELST 507
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP K +G IETW+ AE+ L++AL+K
Sbjct 508 RPEKYVGKIETWDAAESKLESALKK 532
> dre:449661 tars, ThrRS, wu:fb07c05, wu:fb39c12, zgc:92586; threonyl-tRNA
synthetase (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=624
Score = 114 bits (285), Expect = 8e-26, Method: Composition-based stats.
Identities = 51/83 (61%), Positives = 65/83 (78%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFC+++Q+E E L+FL VY+ FGF+F L LST
Sbjct 342 HRNELSGALTGLTRVRRFQQDDAHIFCSMDQIESEIKGCLDFLRTVYDVFGFTFKLNLST 401
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP K +G E W+QAE L+N+L
Sbjct 402 RPEKFLGEPEVWDQAEKQLENSL 424
> mmu:272396 Tarsl2, A530046H20Rik, MGC31414; threonyl-tRNA synthetase-like
2 (EC:6.1.1.3); K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=790
Score = 114 bits (284), Expect = 9e-26, Method: Composition-based stats.
Identities = 55/83 (66%), Positives = 63/83 (75%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+LSGLTRVRRFQQDDAHIFC +EQ+EEE L FL VY FGFSF L LST
Sbjct 508 HRNELSGTLSGLTRVRRFQQDDAHIFCMVEQIEEEIKGCLHFLQSVYSTFGFSFQLNLST 567
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP +G IE W++AE LQN+L
Sbjct 568 RPEHFLGEIEIWDEAERQLQNSL 590
> cpv:cgd7_1710 threonyl-tRNA synthetase (RNA binding domain TGS+HxxxH+tRNA
synthetase) ; K01868 threonyl-tRNA synthetase
[EC:6.1.1.3]
Length=763
Score = 113 bits (282), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 53/85 (62%), Positives = 67/85 (78%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L+GLTRVRRFQQDDAHIFCT EQ++EE AL+FLF++Y Q GF+F L LST
Sbjct 454 HRNEFSGALNGLTRVRRFQQDDAHIFCTPEQIQEEVFKALDFLFFIYGQLGFTFDLFLST 513
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
P + +G E W +AE AL++AL K
Sbjct 514 MPKEHLGTEEQWKEAENALKSALDK 538
> tgo:TGME49_100260 threonyl-tRNA synthetase, putative (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=844
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 53/73 (72%), Positives = 61/73 (83%), Gaps = 0/73 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SGSL+GLTRVRRFQQDDAHIFC L+QV+EE A AL FLF+VY+QFGFSF L LST
Sbjct 516 HRNELSGSLTGLTRVRRFQQDDAHIFCRLDQVKEEVADALNFLFFVYDQFGFSFELFLST 575
Query 61 RPLKAIGCIETWN 73
RP KA+G W+
Sbjct 576 RPKKALGERAVWD 588
> bbo:BBOV_IV008180 23.m05860; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=736
Score = 110 bits (275), Expect = 1e-24, Method: Composition-based stats.
Identities = 52/85 (61%), Positives = 63/85 (74%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E +GSLSGLTRVRRFQQDDAHIFCT EQ+ E S L+F+ VY F F ++ LST
Sbjct 445 HRNELTGSLSGLTRVRRFQQDDAHIFCTPEQISAEVLSMLKFIDDVYSLFNFEYNFKLST 504
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
+P KA+G E WN+AE LQ+AL K
Sbjct 505 KPAKALGDDELWNKAEAGLQDALEK 529
> cel:C47D12.6 trs-1; Threonyl tRNA Synthetase family member (trs-1);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=725
Score = 108 bits (271), Expect = 3e-24, Method: Composition-based stats.
Identities = 53/84 (63%), Positives = 64/84 (76%), Gaps = 1/84 (1%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQ-FGFSFSLALS 59
HR+E SG+L+GLTRVRRFQQDDAHIFC +Q+ EE L+FL Y YE+ FGF+F L LS
Sbjct 440 HRNEMSGALTGLTRVRRFQQDDAHIFCRQDQISEEIKQCLDFLEYAYEKVFGFTFKLNLS 499
Query 60 TRPLKAIGCIETWNQAETALQNAL 83
TRP +G IETW++AE L NAL
Sbjct 500 TRPEGFLGNIETWDKAEADLTNAL 523
> ath:AT1G17960 threonyl-tRNA synthetase, putative / threonine--tRNA
ligase, putative; K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=458
Score = 108 bits (269), Expect = 5e-24, Method: Composition-based stats.
Identities = 50/83 (60%), Positives = 62/83 (74%), Gaps = 0/83 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E SG+L G+TRVRRF QDDAHIFC ++QVEEE L+F+ YVY FGF++ L LST
Sbjct 181 HRNEDSGALGGMTRVRRFVQDDAHIFCRVDQVEEEVKGVLDFIDYVYRIFGFTYELTLST 240
Query 61 RPLKAIGCIETWNQAETALQNAL 83
RP IG +ETW +AE L+ AL
Sbjct 241 RPKDHIGDLETWAKAENDLEKAL 263
> tpv:TP01_0957 threonyl-tRNA synthetase; K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=779
Score = 103 bits (257), Expect = 1e-22, Method: Composition-based stats.
Identities = 50/85 (58%), Positives = 60/85 (70%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+E +GSLSGLTRVRRFQQDDAHIFCT EQ+ EE + L F+ VY F F + LST
Sbjct 498 HRNELTGSLSGLTRVRRFQQDDAHIFCTREQIMEEVLNILTFIGKVYSLFDFRYEFKLST 557
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
+P KA+G E W AE +L+ AL K
Sbjct 558 KPAKALGDNELWEVAEKSLEEALNK 582
> dre:100333765 Threonyl-tRNA synthetase, cytoplasmic-like
Length=529
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 48/84 (57%), Positives = 62/84 (73%), Gaps = 0/84 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR+EASG+L GLTRVRRF QDDAHIFCT EQ+E+E + L+F+ VY FGFSF LST
Sbjct 250 HRNEASGTLGGLTRVRRFCQDDAHIFCTPEQLEQEITACLDFVRSVYRVFGFSFHCLLST 309
Query 61 RPLKAIGCIETWNQAETALQNALR 84
RP ++G W+ AE L+++L+
Sbjct 310 RPASSMGDSALWDCAEQQLESSLK 333
> hsa:80222 TARS2, FLJ12528, TARSL1; threonyl-tRNA synthetase
2, mitochondrial (putative) (EC:6.1.1.3); K01868 threonyl-tRNA
synthetase [EC:6.1.1.3]
Length=718
Score = 98.6 bits (244), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 51/85 (60%), Positives = 60/85 (70%), Gaps = 0/85 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR EASG L GLTR+R FQQDDAHIFCT +Q+E E S L+FL VY GFSF LALST
Sbjct 437 HRAEASGGLGGLTRLRCFQQDDAHIFCTTDQLEAEIQSCLDFLRSVYAVLGFSFRLALST 496
Query 61 RPLKAIGCIETWNQAETALQNALRK 85
RP +G W+QAE L+ AL++
Sbjct 497 RPSGFLGDPCLWDQAEQVLKQALKE 521
> mmu:71807 Tars2, 2610024N01Rik, AI429208, Tarsl1; threonyl-tRNA
synthetase 2, mitochondrial (putative) (EC:6.1.1.3); K01868
threonyl-tRNA synthetase [EC:6.1.1.3]
Length=697
Score = 91.7 bits (226), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 47/76 (61%), Positives = 52/76 (68%), Gaps = 0/76 (0%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALST 60
HR EASGSL GLTR+ RFQQDDAHIFC Q+E E L+FL VY GFSF LALST
Sbjct 442 HRAEASGSLGGLTRLWRFQQDDAHIFCAPHQLEAEIQGCLDFLRCVYSVLGFSFHLALST 501
Query 61 RPLKAIGCIETWNQAE 76
RP +G W+QAE
Sbjct 502 RPPGFLGEPRLWDQAE 517
> eco:b1719 thrS, ECK1717, JW1709; threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=642
Score = 87.4 bits (215), Expect = 1e-17, Method: Composition-based stats.
Identities = 44/87 (50%), Positives = 56/87 (64%), Gaps = 1/87 (1%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFS-FSLALS 59
HR+E SGSL GL RVR F QDDAHIFCT EQ+ +E + ++ +Y FGF + LS
Sbjct 362 HRNEPSGSLHGLMRVRGFTQDDAHIFCTEEQIRDEVNGCIRLVYDMYSTFGFEKIVVKLS 421
Query 60 TRPLKAIGCIETWNQAETALQNALRKH 86
TRP K IG E W++AE L AL ++
Sbjct 422 TRPEKRIGSDEMWDRAEADLAVALEEN 448
> sce:YKL194C MST1; Mitochondrial threonyl-tRNA synthetase (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=462
Score = 80.1 bits (196), Expect = 2e-15, Method: Composition-based stats.
Identities = 42/95 (44%), Positives = 59/95 (62%), Gaps = 10/95 (10%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQ-FGF------- 52
HR+EASG+LSGLTR+R+F QDD HIFCT QV+ E ++L+ + VY + F F
Sbjct 161 HRNEASGALSGLTRLRKFHQDDGHIFCTPSQVKSEIFNSLKLIDIVYNKIFPFVKGGSGA 220
Query 53 --SFSLALSTRPLKAIGCIETWNQAETALQNALRK 85
++ + STRP IG ++ WN AE L+ L +
Sbjct 221 ESNYFINFSTRPDHFIGDLKVWNHAEQVLKEILEE 255
> ath:AT2G04842 EMB2761 (EMBRYO DEFECTIVE 2761); ATP binding /
aminoacyl-tRNA ligase/ ligase, forming aminoacyl-tRNA and related
compounds / nucleotide binding / threonine-tRNA ligase
(EC:6.1.1.3); K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=650
Score = 77.8 bits (190), Expect = 8e-15, Method: Composition-based stats.
Identities = 39/84 (46%), Positives = 56/84 (66%), Gaps = 1/84 (1%)
Query 1 HRDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFS-FSLALS 59
+R E SGSL GL RVR F QDDAHIFC +Q+++E L+ + +FGF+ + + LS
Sbjct 375 YRYELSGSLHGLFRVRGFTQDDAHIFCLEDQIKDEIRGVLDLTEEILSRFGFNKYEVNLS 434
Query 60 TRPLKAIGCIETWNQAETALQNAL 83
TRP K++G + W +A AL++AL
Sbjct 435 TRPEKSVGGDDIWEKATCALRDAL 458
> dre:100330276 threonyl-tRNA synthetase-like
Length=380
Score = 54.3 bits (129), Expect = 9e-08, Method: Composition-based stats.
Identities = 27/55 (49%), Positives = 34/55 (61%), Gaps = 0/55 (0%)
Query 31 QVEEEGASALEFLFYVYEQFGFSFSLALSTRPLKAIGCIETWNQAETALQNALRK 85
++ E L+FL VY FGFSF L LSTRP K +G WNQAE L+N+L +
Sbjct 128 KIGSEMKGCLDFLRSVYSVFGFSFQLHLSTRPQKFLGDEALWNQAEKQLENSLNE 182
> tgo:TGME49_019430 threonyl-tRNA synthetase, putative (EC:6.1.1.3);
K01868 threonyl-tRNA synthetase [EC:6.1.1.3]
Length=1223
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 47/97 (48%), Gaps = 13/97 (13%)
Query 2 RDEASGSLSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQ-FGFS---FSLA 57
R E +L GL R+R F QDD HI CT EQ EE L + ++Y Q FG S ++
Sbjct 922 RRERRSALCGLFRLREFTQDDGHILCTREQAVEEIKVQLASILHLYVQVFGISPEGVKVS 981
Query 58 LSTRPLKAIGCIET---------WNQAETALQNALRK 85
L +RP KA + W QAE L A R+
Sbjct 982 LGSRPAKASRRSDASADSEDEKLWTQAEAWLLEAARE 1018
> ath:AT3G60970 ATMRP15; ATPase, coupled to transmembrane movement
of substances
Length=1037
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 9/60 (15%)
Query 8 SLSGLTRVRRFQQDDAHIFCTLEQVEEEG------ASALEFLFY---VYEQFGFSFSLAL 58
SL+G T +R F Q D I L ++ ASA+E+L + + F F+FSL L
Sbjct 650 SLAGATTIRAFDQRDRFISSNLVLIDSHSRPWFHVASAMEWLSFRLNLLSHFVFAFSLVL 709
> tpv:TP03_0720 hypothetical protein
Length=575
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 29/61 (47%), Gaps = 15/61 (24%)
Query 9 LSGLTRVRRFQQDDAHIFCTLEQVEEEGASALEFLFYVYEQFGFSFSLALSTRPLKAIGC 68
LSGL R++RF DDA+ F + L +Y GF+ +L + P+ + C
Sbjct 56 LSGLFRIQRFFSDDAYYFS-------------KQLTLMYGVIGFTLTLLIG--PIGKVAC 100
Query 69 I 69
I
Sbjct 101 I 101
> hsa:5257 PHKB, DKFZp781E15103, FLJ41698; phosphorylase kinase,
beta (EC:2.7.11.19 2.7.1.38); K07190 phosphorylase kinase
alpha/beta subunit
Length=1093
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 28 TLEQVEEEGASALEFLFYVYEQFGFSFSLALSTRPLKAIGCIET 71
T +QVE + L Y Q G + L LS RP + IGC+ T
Sbjct 519 TPQQVEPIQIWPQQELVKAYLQLGINEKLGLSGRPDRPIGCLGT 562
> mmu:226432 Ipo9, 0710008K06Rik, AI845704, C78347, Imp9, Ranbp9,
mKIAA1192; importin 9
Length=1040
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 4/67 (5%)
Query 23 AHIFCTLEQVEEEGASALEFLFYVYEQFGFSF--SLALSTRPLKAIGCIETWNQAETALQ 80
AH+ C +E++E+ A L +F V +QF +F +L + P G +A TAL
Sbjct 219 AHMICNMEELEKGAAKVL--IFPVVQQFTEAFVQALQMPDGPTSDSGFKMEVLKAVTALV 276
Query 81 NALRKHI 87
KH+
Sbjct 277 KNFPKHM 283
> ath:AT3G60160 ATMRP9; ATMRP9; ATPase, coupled to transmembrane
movement of substances
Length=1490
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 30/60 (50%), Gaps = 9/60 (15%)
Query 8 SLSGLTRVRRFQQDDAHIFCTLEQVEEEG------ASALEFLFY---VYEQFGFSFSLAL 58
SL+G T +R F Q D I L ++ ASA+E+L + + F F+FSL L
Sbjct 1103 SLAGATTIRAFDQRDRFISSNLVLIDSHSRPWFHVASAMEWLSFRLNLLSHFVFAFSLVL 1162
> hsa:55705 IPO9, DKFZp761M1547, FLJ10402, Imp9; importin 9
Length=1041
Score = 28.9 bits (63), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 33/67 (49%), Gaps = 4/67 (5%)
Query 23 AHIFCTLEQVEEEGASALEFLFYVYEQFGFSF--SLALSTRPLKAIGCIETWNQAETALQ 80
AH+ C +E++E+ A L +F V +QF +F +L + P G +A TAL
Sbjct 219 AHMICNMEELEKGAAKVL--IFPVVQQFTEAFVQALQIPDGPTSDSGFKMEVLKAVTALV 276
Query 81 NALRKHI 87
KH+
Sbjct 277 KNFPKHM 283
> hsa:683 BST1, CD157; bone marrow stromal cell antigen 1 (EC:3.2.2.5);
K01242 NAD+ nucleosidase [EC:3.2.2.5]
Length=318
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 14/51 (27%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 32 VEEEGASALEFLFYVYEQFGFSFSLALSTRPLKAIGCIETWNQAETALQNA 82
VE G +++ L + GF +S RP+K + C++ + AL++A
Sbjct 235 VESCGEGSMKVLEKRLKDMGFQYSCINDYRPVKLLQCVDHSTHPDCALKSA 285
Lambda K H
0.322 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2026251472
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40