bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2587_orf1
Length=67
Score E
Sequences producing significant alignments: (Bits) Value
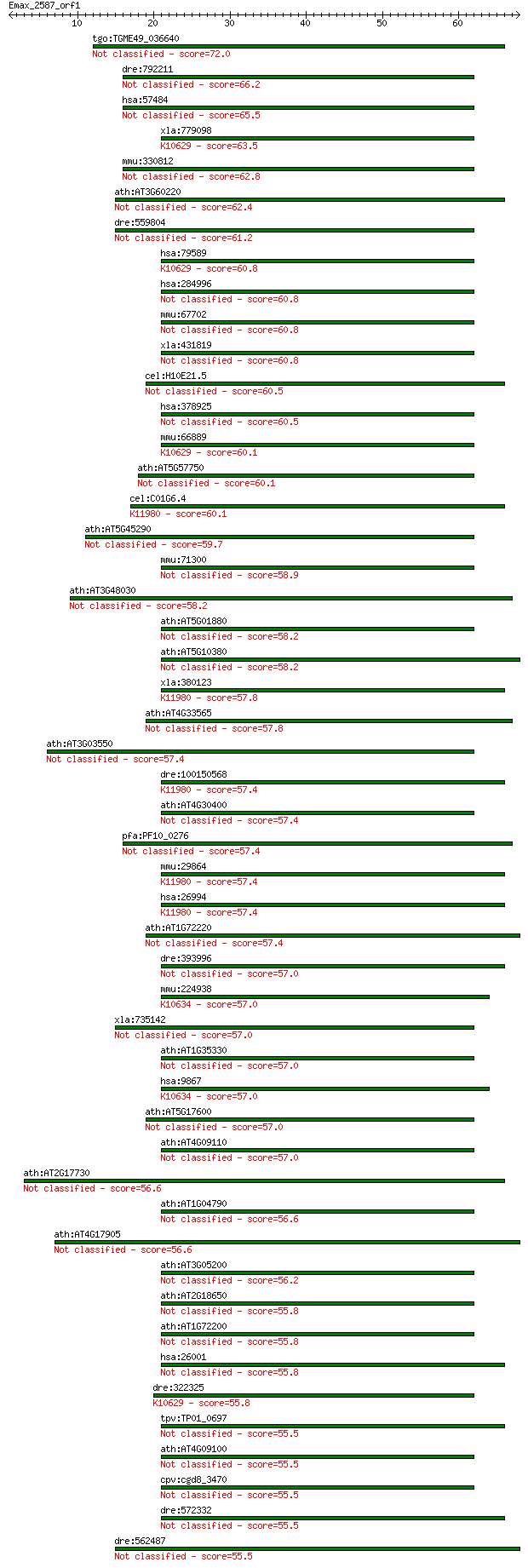
tgo:TGME49_036640 zinc finger (C3HC4 RING finger) protein, put... 72.0 4e-13
dre:792211 rnf150, MGC92168, fi49f09, wu:fi49f09, zgc:92168; r... 66.2 2e-11
hsa:57484 RNF150, MGC125502; ring finger protein 150 65.5
xla:779098 rnf128, grail, greul1; ring finger protein 128; K10... 63.5 1e-10
mmu:330812 Rnf150, A630007N06Rik, C030044C12Rik, Greul5, KIAA1... 62.8 3e-10
ath:AT3G60220 ATL4; ATL4; protein binding / zinc ion binding 62.4 3e-10
dre:559804 similar to RING finger protein 150 61.2
hsa:79589 RNF128, FLJ23516, GRAIL; ring finger protein 128 (EC... 60.8 9e-10
hsa:284996 RNF149, DNAPTP2, FLJ90504; ring finger protein 149 60.8
mmu:67702 Rnf149, 1600023E10Rik, AV037261, C76763, Gm15832, Gr... 60.8 1e-09
xla:431819 rnf149, MGC81168; ring finger protein 149 60.8
cel:H10E21.5 hypothetical protein 60.5 1e-09
hsa:378925 RNF148, MGC35222; ring finger protein 148 60.5
mmu:66889 Rnf128, 1300002C13Rik, AI987883, GRAIL, Greul1; ring... 60.1 2e-09
ath:AT5G57750 zinc finger (C3HC4-type RING finger) family protein 60.1 2e-09
cel:C01G6.4 hypothetical protein; K11980 E3 ubiquitin-protein ... 60.1 2e-09
ath:AT5G45290 zinc finger (C3HC4-type RING finger) family protein 59.7 2e-09
mmu:71300 Rnf148, 4933432M07Rik, Greul3; ring finger protein 148 58.9
ath:AT3G48030 hypoxia-responsive family protein / zinc finger ... 58.2 6e-09
ath:AT5G01880 zinc finger (C3HC4-type RING finger) family protein 58.2 7e-09
ath:AT5G10380 RING1; RING1; protein binding / ubiquitin-protei... 58.2 7e-09
xla:380123 rnf11, MGC130820, MGC52858, rnf11-a; ring finger pr... 57.8 8e-09
ath:AT4G33565 protein binding / zinc ion binding 57.8 8e-09
ath:AT3G03550 zinc finger (C3HC4-type RING finger) family protein 57.4 1e-08
dre:100150568 MGC114095; zgc:114095; K11980 E3 ubiquitin-prote... 57.4 1e-08
ath:AT4G30400 zinc finger (C3HC4-type RING finger) family protein 57.4 1e-08
pfa:PF10_0276 Zinc finger, C3HC4 type, putative 57.4 1e-08
mmu:29864 Rnf11; ring finger protein 11; K11980 E3 ubiquitin-p... 57.4 1e-08
hsa:26994 RNF11, MGC51169, SID1669; ring finger protein 11; K1... 57.4 1e-08
ath:AT1G72220 zinc finger (C3HC4-type RING finger) family protein 57.4 1e-08
dre:393996 rnf11, MGC63861, zgc:63861; ring finger protein 11 57.0
mmu:224938 Pja2, AI447901, AL022700, MGC27629, Neurodap1, mKIA... 57.0 1e-08
xla:735142 znrf3, MGC115570; zinc and ring finger 3 57.0
ath:AT1G35330 zinc finger (C3HC4-type RING finger) family protein 57.0 1e-08
hsa:9867 PJA2, KIAA0438, Neurodap1, RNF131; praja ring finger ... 57.0 1e-08
ath:AT5G17600 zinc finger (C3HC4-type RING finger) family protein 57.0 2e-08
ath:AT4G09110 zinc finger (C3HC4-type RING finger) family protein 57.0 2e-08
ath:AT2G17730 zinc finger (C3HC4-type RING finger) family protein 56.6 2e-08
ath:AT1G04790 zinc finger (C3HC4-type RING finger) family protein 56.6 2e-08
ath:AT4G17905 ATL4H; ATL4H; protein binding / zinc ion binding 56.6 2e-08
ath:AT3G05200 ATL6; ATL6; protein binding / zinc ion binding 56.2 3e-08
ath:AT2G18650 MEE16; MEE16 (maternal effect embryo arrest 16);... 55.8 3e-08
ath:AT1G72200 zinc finger (C3HC4-type RING finger) family protein 55.8 3e-08
hsa:26001 RNF167, 5730408C10Rik, DKFZp566H073, RING105; ring f... 55.8 3e-08
dre:322325 rnf128a, rnf128, wu:fb57h05, zgc:77843; ring finger... 55.8 3e-08
tpv:TP01_0697 hypothetical protein 55.5 4e-08
ath:AT4G09100 zinc finger (C3HC4-type RING finger) family protein 55.5 4e-08
cpv:cgd8_3470 membrane associated protein with a RING finger, ... 55.5 4e-08
dre:572332 ring finger protein 133-like 55.5 4e-08
dre:562487 XRnf12-like 55.5 4e-08
> tgo:TGME49_036640 zinc finger (C3HC4 RING finger) protein, putative
Length=511
Score = 72.0 bits (175), Expect = 4e-13, Method: Composition-based stats.
Identities = 30/54 (55%), Positives = 34/54 (62%), Gaps = 0/54 (0%)
Query 12 GAFSSTFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
FS C ICLGEF E+ IR C HFFH CID WL+K++ CPLCLR I
Sbjct 442 ATFSPVQENCCICLGEFADEEVIRELKCSHFFHHGCIDKWLLKNKQCPLCLRSI 495
> dre:792211 rnf150, MGC92168, fi49f09, wu:fi49f09, zgc:92168;
ring finger protein 150
Length=419
Score = 66.2 bits (160), Expect = 2e-11, Method: Composition-based stats.
Identities = 22/46 (47%), Positives = 30/46 (65%), Gaps = 0/46 (0%)
Query 16 STFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
S F C +C+ + P D +R+ PC+H FHK C+D WLV R CP+C
Sbjct 262 SDFDNCAVCIEGYKPNDVVRILPCRHLFHKCCVDPWLVDHRTCPMC 307
> hsa:57484 RNF150, MGC125502; ring finger protein 150
Length=438
Score = 65.5 bits (158), Expect = 4e-11, Method: Composition-based stats.
Identities = 21/46 (45%), Positives = 31/46 (67%), Gaps = 0/46 (0%)
Query 16 STFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
S F C +C+ + P D +R+ PC+H FHK+C+D WL+ R CP+C
Sbjct 273 SDFDNCAVCIEGYKPNDVVRILPCRHLFHKSCVDPWLLDHRTCPMC 318
> xla:779098 rnf128, grail, greul1; ring finger protein 128; K10629
E3 ubiquitin-protein ligase RNF128 [EC:6.3.2.19]
Length=404
Score = 63.5 bits (153), Expect = 1e-10, Method: Composition-based stats.
Identities = 20/41 (48%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
C +C+ + P D +R+ C HFFHK CID WL++ R CP+C
Sbjct 260 CAVCIEPYKPSDVVRILTCNHFFHKNCIDPWLLEHRTCPMC 300
> mmu:330812 Rnf150, A630007N06Rik, C030044C12Rik, Greul5, KIAA1214,
mKIAA1214; ring finger protein 150
Length=437
Score = 62.8 bits (151), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 31/46 (67%), Gaps = 0/46 (0%)
Query 16 STFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
S F C +C+ + P D +R+ PC+H FHK+C+D WL+ R CP+C
Sbjct 272 SDFDNCAVCIEGYKPNDVVRILPCRHLFHKSCVDPWLLDHRTCPMC 317
> ath:AT3G60220 ATL4; ATL4; protein binding / zinc ion binding
Length=334
Score = 62.4 bits (150), Expect = 3e-10, Method: Composition-based stats.
Identities = 29/53 (54%), Positives = 35/53 (66%), Gaps = 2/53 (3%)
Query 15 SSTFSG-CVICLGEFVPEDTIRLPP-CQHFFHKTCIDAWLVKSRNCPLCLRPI 65
SS SG C +CL +F PED +RL P C H FH CID WLV ++ CPLC P+
Sbjct 110 SSMNSGDCAVCLSKFEPEDQLRLLPLCCHAFHADCIDIWLVSNQTCPLCRSPL 162
> dre:559804 similar to RING finger protein 150
Length=418
Score = 61.2 bits (147), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 20/47 (42%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 15 SSTFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
S F C +C+ ++ P D +R+ PC+H FH+ C+D WL R CP+C
Sbjct 259 DSDFDNCAVCIEDYKPNDVVRILPCRHVFHRNCVDPWLQDHRTCPMC 305
> hsa:79589 RNF128, FLJ23516, GRAIL; ring finger protein 128 (EC:6.3.2.-);
K10629 E3 ubiquitin-protein ligase RNF128 [EC:6.3.2.19]
Length=402
Score = 60.8 bits (146), Expect = 9e-10, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
C +C+ + P D +R+ C H FHKTC+D WL++ R CP+C
Sbjct 251 CAVCIELYKPNDLVRILTCNHIFHKTCVDPWLLEHRTCPMC 291
> hsa:284996 RNF149, DNAPTP2, FLJ90504; ring finger protein 149
Length=400
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 20/41 (48%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
C +C+ F +D IR+ PC+H FH+ CID WL+ R CP+C
Sbjct 269 CAVCIENFKVKDIIRILPCKHIFHRICIDPWLLDHRTCPMC 309
> mmu:67702 Rnf149, 1600023E10Rik, AV037261, C76763, Gm15832,
Greul4, OTTMUSG00000026591; ring finger protein 149
Length=394
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 20/41 (48%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
C +C+ F +D IR+ PC+H FH+ CID WL+ R CP+C
Sbjct 265 CAVCIENFKVKDVIRILPCKHIFHRICIDPWLLDHRTCPMC 305
> xla:431819 rnf149, MGC81168; ring finger protein 149
Length=397
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 29/41 (70%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
C +C+ + +D +R+ PC+H FH+ CID WL++ R CP+C
Sbjct 264 CAVCIENYKTKDLVRILPCKHIFHRLCIDPWLIEHRTCPMC 304
> cel:H10E21.5 hypothetical protein
Length=473
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 23/47 (48%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 19 SGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
S C +CL + +D IRL PC+H +HK+CID WL++ R CP+C I
Sbjct 225 SDCAVCLDPYQLQDVIRLLPCKHIYHKSCIDPWLLEHRTCPMCKNDI 271
> hsa:378925 RNF148, MGC35222; ring finger protein 148
Length=305
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 21/41 (51%), Positives = 29/41 (70%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
CV+C + P+D +R+ C+HFFHK CID WL+ R CP+C
Sbjct 258 CVVCFDTYKPQDVVRILTCKHFFHKACIDPWLLAHRTCPMC 298
> mmu:66889 Rnf128, 1300002C13Rik, AI987883, GRAIL, Greul1; ring
finger protein 128 (EC:6.3.2.-); K10629 E3 ubiquitin-protein
ligase RNF128 [EC:6.3.2.19]
Length=428
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
C +C+ + P D +R+ C H FHKTC+D WL++ R CP+C
Sbjct 277 CAVCIELYKPNDLVRILTCNHIFHKTCVDPWLLEHRTCPMC 317
> ath:AT5G57750 zinc finger (C3HC4-type RING finger) family protein
Length=210
Score = 60.1 bits (144), Expect = 2e-09, Method: Composition-based stats.
Identities = 24/45 (53%), Positives = 28/45 (62%), Gaps = 1/45 (2%)
Query 18 FSGCVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
S C +CL EF ED +RL P C H FH CID WL+ + CPLC
Sbjct 119 LSDCAVCLREFTAEDELRLLPKCSHAFHVECIDTWLLTNSTCPLC 163
> cel:C01G6.4 hypothetical protein; K11980 E3 ubiquitin-protein
ligase RNF11
Length=170
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 32/49 (65%), Gaps = 0/49 (0%)
Query 17 TFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
T + C IC+ +F P + IR PC H FH+ C+D WL+KS CP CL P+
Sbjct 91 TSNECAICMIDFEPGERIRFLPCMHSFHQECVDEWLMKSFTCPSCLEPV 139
> ath:AT5G45290 zinc finger (C3HC4-type RING finger) family protein
Length=546
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 28/53 (52%), Positives = 33/53 (62%), Gaps = 2/53 (3%)
Query 11 TGAFSSTFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWL--VKSRNCPLC 61
T + S S C ICL E+ D+IR PC H FHKTC+D WL + SR CPLC
Sbjct 480 TKSQSEDPSQCYICLVEYEEADSIRTLPCHHEFHKTCVDKWLKEIHSRVCPLC 532
> mmu:71300 Rnf148, 4933432M07Rik, Greul3; ring finger protein
148
Length=316
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 22/41 (53%), Positives = 29/41 (70%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
CV+C + +D IR+ C+HFFHKTCID WL+ R CP+C
Sbjct 269 CVVCFDMYKAQDVIRILTCKHFFHKTCIDPWLLAHRTCPMC 309
> ath:AT3G48030 hypoxia-responsive family protein / zinc finger
(C3HC4-type RING finger) family protein
Length=349
Score = 58.2 bits (139), Expect = 6e-09, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 34/59 (57%), Gaps = 2/59 (3%)
Query 9 NMTGAFSSTFSGCVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLCLRPIA 66
N+T + F C +CL EF D +RL P C H FH CID WL+ + CPLC R ++
Sbjct 196 NVTISLEQPFD-CAVCLNEFSDTDKLRLLPVCSHAFHLHCIDTWLLSNSTCPLCRRSLS 253
> ath:AT5G01880 zinc finger (C3HC4-type RING finger) family protein
Length=159
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 24/42 (57%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 21 CVICLGEFVPEDTIR-LPPCQHFFHKTCIDAWLVKSRNCPLC 61
C ICLGEF + +R LPPC H FH +CID WLV +CP C
Sbjct 105 CAICLGEFADGERVRVLPPCNHSFHMSCIDTWLVSHSSCPNC 146
> ath:AT5G10380 RING1; RING1; protein binding / ubiquitin-protein
ligase/ zinc ion binding
Length=301
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 23/48 (47%), Positives = 32/48 (66%), Gaps = 1/48 (2%)
Query 21 CVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLCLRPIAI 67
C +CL EF ++++RL P C H FH CID WL+ +NCPLC P+ +
Sbjct 135 CSVCLNEFEEDESLRLLPKCSHAFHLNCIDTWLLSHKNCPLCRAPVLL 182
> xla:380123 rnf11, MGC130820, MGC52858, rnf11-a; ring finger
protein 11; K11980 E3 ubiquitin-protein ligase RNF11
Length=154
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 23/45 (51%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
CVIC+ +FV D IR PC H +H CID WL++S CP C+ P+
Sbjct 99 CVICMMDFVYGDPIRFLPCMHIYHMDCIDDWLMRSFTCPSCMEPV 143
> ath:AT4G33565 protein binding / zinc ion binding
Length=367
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 26/49 (53%), Positives = 31/49 (63%), Gaps = 1/49 (2%)
Query 19 SGCVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLCLRPIA 66
+ C +CL EF E+T+RL P C+H FH CID WL NCPLC PI
Sbjct 215 TDCSVCLSEFEEEETLRLLPKCKHAFHLYCIDTWLRSHTNCPLCRAPIV 263
> ath:AT3G03550 zinc finger (C3HC4-type RING finger) family protein
Length=356
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/57 (47%), Positives = 32/57 (56%), Gaps = 3/57 (5%)
Query 6 RKRNMTGAFSSTFSGCVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
+ R M G S S C +CL EF +++RL P C H FH CID WL NCPLC
Sbjct 146 KYRKMDGFVES--SDCSVCLSEFQENESLRLLPKCNHAFHVPCIDTWLKSHSNCPLC 200
> dre:100150568 MGC114095; zgc:114095; K11980 E3 ubiquitin-protein
ligase RNF11
Length=154
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/45 (51%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
CVIC+ +FV D IR PC H +H CID WL++S CP C+ P+
Sbjct 99 CVICMMDFVYGDPIRFLPCMHIYHLDCIDDWLMRSFTCPSCMEPV 143
> ath:AT4G30400 zinc finger (C3HC4-type RING finger) family protein
Length=472
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 21 CVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
C +CL EF ED +RL P C H FH CID WL+ CPLC
Sbjct 134 CAVCLCEFETEDKLRLLPKCSHAFHMDCIDTWLLSHSTCPLC 175
> pfa:PF10_0276 Zinc finger, C3HC4 type, putative
Length=274
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/52 (50%), Positives = 33/52 (63%), Gaps = 1/52 (1%)
Query 16 STFSGCVICLGEFVPEDTIR-LPPCQHFFHKTCIDAWLVKSRNCPLCLRPIA 66
S S C ICL +F ++ +R L C H FHK+CID WL++S CP C PIA
Sbjct 215 SNESKCSICLNDFQIDECVRTLLLCNHTFHKSCIDLWLIRSATCPNCKSPIA 266
> mmu:29864 Rnf11; ring finger protein 11; K11980 E3 ubiquitin-protein
ligase RNF11
Length=154
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/45 (51%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
CVIC+ +FV D IR PC H +H CID WL++S CP C+ P+
Sbjct 99 CVICMMDFVYGDPIRFLPCMHIYHLDCIDDWLMRSFTCPSCMEPV 143
> hsa:26994 RNF11, MGC51169, SID1669; ring finger protein 11;
K11980 E3 ubiquitin-protein ligase RNF11
Length=154
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/45 (51%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
CVIC+ +FV D IR PC H +H CID WL++S CP C+ P+
Sbjct 99 CVICMMDFVYGDPIRFLPCMHIYHLDCIDDWLMRSFTCPSCMEPV 143
> ath:AT1G72220 zinc finger (C3HC4-type RING finger) family protein
Length=413
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 24/50 (48%), Positives = 32/50 (64%), Gaps = 1/50 (2%)
Query 19 SGCVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLCLRPIAI 67
+ C +CL EF ++++RL P C H FH +CID WL NCPLC IA+
Sbjct 175 TDCPVCLNEFEEDESLRLLPKCNHAFHISCIDTWLSSHTNCPLCRAGIAM 224
> dre:393996 rnf11, MGC63861, zgc:63861; ring finger protein 11
Length=146
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/45 (51%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
CVIC+ +F D IR PC H +H CIDAWL++S CP C+ P+
Sbjct 91 CVICMMDFEYGDPIRFLPCMHIYHVDCIDAWLMRSFTCPSCMEPV 135
> mmu:224938 Pja2, AI447901, AL022700, MGC27629, Neurodap1, mKIAA0438;
praja 2, RING-H2 motif containing; K10634 E3 ubiquitin-protein
ligase Praja2 [EC:6.3.2.19]
Length=707
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 21/43 (48%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLR 63
C IC E++ +D PC HFFHK C+ WL KS CP+C R
Sbjct 633 CPICCSEYIKDDIATELPCHHFFHKPCVSIWLQKSGTCPVCRR 675
> xla:735142 znrf3, MGC115570; zinc and ring finger 3
Length=784
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 21/47 (44%), Positives = 32/47 (68%), Gaps = 0/47 (0%)
Query 15 SSTFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
SS+ S C ICL +++ + +R+ PC H FHK C+D WL+++ CP C
Sbjct 196 SSSISDCAICLEKYIDGEELRVIPCTHRFHKRCVDPWLLQNHTCPHC 242
> ath:AT1G35330 zinc finger (C3HC4-type RING finger) family protein
Length=327
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 24/42 (57%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 21 CVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
C ICL EF E+T+RL PPC H FH +CID WL CP+C
Sbjct 128 CAICLNEFEDEETLRLMPPCSHAFHASCIDVWLSSRSTCPVC 169
> hsa:9867 PJA2, KIAA0438, Neurodap1, RNF131; praja ring finger
2 (EC:6.3.2.-); K10634 E3 ubiquitin-protein ligase Praja2
[EC:6.3.2.19]
Length=708
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 21/43 (48%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLR 63
C IC E++ +D PC HFFHK C+ WL KS CP+C R
Sbjct 634 CPICCSEYIKDDIATELPCHHFFHKPCVSIWLQKSGTCPVCRR 676
> ath:AT5G17600 zinc finger (C3HC4-type RING finger) family protein
Length=362
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 23/44 (52%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 19 SGCVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
S C +CL EF +++RL P C H FH CID WL NCPLC
Sbjct 140 SDCSVCLSEFEENESLRLLPKCNHAFHLPCIDTWLKSHSNCPLC 183
> ath:AT4G09110 zinc finger (C3HC4-type RING finger) family protein
Length=302
Score = 57.0 bits (136), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/42 (54%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 21 CVICLGEFVPEDTIR-LPPCQHFFHKTCIDAWLVKSRNCPLC 61
C ICL EFV ++T+R +PPC H FH CID WL CP C
Sbjct 123 CAICLSEFVDKETLRWMPPCSHTFHANCIDVWLSSQSTCPAC 164
> ath:AT2G17730 zinc finger (C3HC4-type RING finger) family protein
Length=241
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 28/68 (41%), Positives = 39/68 (57%), Gaps = 5/68 (7%)
Query 3 EKQRKRNMTGAFSSTFS----GCVICLGEFVPEDTIR-LPPCQHFFHKTCIDAWLVKSRN 57
EK K +TG ++ S C +CL +F +T+R LP C H FH CID WL++ +
Sbjct 174 EKIPKMTITGNNNTDASENTDSCSVCLQDFQLGETVRSLPHCHHMFHLPCIDNWLLRHGS 233
Query 58 CPLCLRPI 65
CP+C R I
Sbjct 234 CPMCRRDI 241
> ath:AT1G04790 zinc finger (C3HC4-type RING finger) family protein
Length=634
Score = 56.6 bits (135), Expect = 2e-08, Method: Composition-based stats.
Identities = 24/41 (58%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
CVICL DTIR PC H FHK CID WL +S++CP+C
Sbjct 589 CVICLETPKIGDTIRHLPCLHKFHKDCIDPWLGRSKSCPVC 629
> ath:AT4G17905 ATL4H; ATL4H; protein binding / zinc ion binding
Length=310
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 28/62 (45%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Query 7 KRNMTGAFSSTFSGCVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
K++ G F + C ICLGEF ++++RL P C H FH CID WL NCPLC I
Sbjct 142 KKHQNG-FKINGTDCSICLGEFNEDESLRLLPKCNHTFHVVCIDRWLKSHSNCPLCRAKI 200
Query 66 AI 67
+
Sbjct 201 IV 202
> ath:AT3G05200 ATL6; ATL6; protein binding / zinc ion binding
Length=398
Score = 56.2 bits (134), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 21 CVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
C ICL EF ++T+RL P C H FH CIDAWL CP+C
Sbjct 128 CAICLNEFEDDETLRLLPKCDHVFHPHCIDAWLEAHVTCPVC 169
> ath:AT2G18650 MEE16; MEE16 (maternal effect embryo arrest 16);
protein binding / zinc ion binding
Length=423
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Query 21 CVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
C +CL EF ED +RL P C H FH CID WL+ CPLC
Sbjct 126 CPVCLCEFETEDKLRLLPKCSHAFHVECIDTWLLSHSTCPLC 167
> ath:AT1G72200 zinc finger (C3HC4-type RING finger) family protein
Length=404
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 23/42 (54%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 21 CVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
C +CL EF ++T+RL P C H FH CIDAWL CPLC
Sbjct 144 CSVCLNEFEDDETLRLIPKCCHVFHPGCIDAWLRSHTTCPLC 185
> hsa:26001 RNF167, 5730408C10Rik, DKFZp566H073, RING105; ring
finger protein 167
Length=350
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 20/46 (43%), Positives = 30/46 (65%), Gaps = 1/46 (2%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSR-NCPLCLRPI 65
C ICL E+ D +R+ PC H +H C+D WL ++R CP+C +P+
Sbjct 230 CAICLDEYEDGDKLRVLPCAHAYHSRCVDPWLTQTRKTCPICKQPV 275
> dre:322325 rnf128a, rnf128, wu:fb57h05, zgc:77843; ring finger
protein 128a (EC:6.3.2.-); K10629 E3 ubiquitin-protein ligase
RNF128 [EC:6.3.2.19]
Length=389
Score = 55.8 bits (133), Expect = 3e-08, Method: Composition-based stats.
Identities = 17/42 (40%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 20 GCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
C +C+ + D + + C HFFHK+CI+ WL++ R CP+C
Sbjct 255 ACAVCIDSYKAGDVLSILTCNHFFHKSCIEPWLLEHRTCPMC 296
> tpv:TP01_0697 hypothetical protein
Length=285
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 22/45 (48%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
C ICL +F ED +R+ C H FH CID WL +S CPLC++ +
Sbjct 240 CGICLDDFAAEDMLRVLHCSHGFHTKCIDLWLSRSVVCPLCMKTL 284
> ath:AT4G09100 zinc finger (C3HC4-type RING finger) family protein
Length=132
Score = 55.5 bits (132), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 23/42 (54%), Positives = 29/42 (69%), Gaps = 1/42 (2%)
Query 21 CVICLGEFVPEDTIRL-PPCQHFFHKTCIDAWLVKSRNCPLC 61
CV+CL EF ++T+RL PPC H FH C+D WL S CP+C
Sbjct 85 CVVCLNEFKDDETLRLVPPCVHVFHADCVDIWLSHSSTCPIC 126
> cpv:cgd8_3470 membrane associated protein with a RING finger,
4xtransmembrane domain
Length=437
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 20/41 (48%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLC 61
C+ICL +F + R PC H FH CID WL+++ CPLC
Sbjct 392 CIICLNDFSSFEMARCLPCNHVFHDDCIDMWLLRNAVCPLC 432
> dre:572332 ring finger protein 133-like
Length=710
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 22/45 (48%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 21 CVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPI 65
C ICL EF+ +R+ C H FHK C+D WL++ R CPLC+ I
Sbjct 146 CAICLEEFMDGQDLRIISCAHEFHKECVDPWLLQHRTCPLCMHNI 190
> dre:562487 XRnf12-like
Length=632
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 20/53 (37%), Positives = 30/53 (56%), Gaps = 0/53 (0%)
Query 15 SSTFSGCVICLGEFVPEDTIRLPPCQHFFHKTCIDAWLVKSRNCPLCLRPIAI 67
S F C +C+ E+ + +R PC H +H CID WL ++ CP+C R + I
Sbjct 572 SDAFKTCSVCITEYAEGNKLRKLPCSHEYHVHCIDRWLSENSTCPICRRAVLI 624
Lambda K H
0.329 0.141 0.484
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2038271796
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40