bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2736_orf1
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
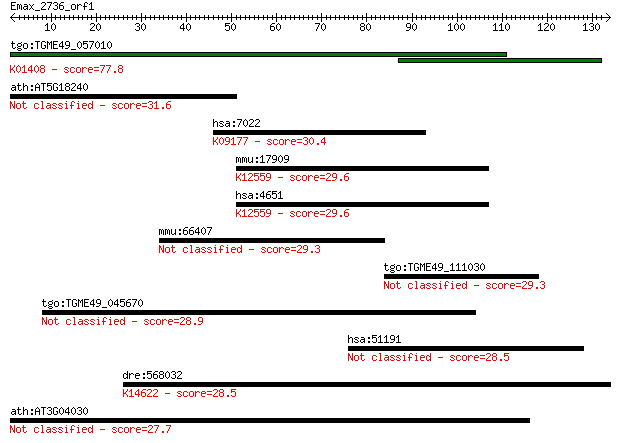
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 77.8 9e-15
ath:AT5G18240 MYR1; MYR1 (MYb-related protein 1); transcriptio... 31.6 0.76
hsa:7022 TFAP2C, AP2-GAMMA, ERF1, TFAP2G, hAP-2g; transcriptio... 30.4 1.5
mmu:17909 Myo10, AW048724, D15Ertd600e, mKIAA0799; myosin X; K... 29.6 2.8
hsa:4651 MYO10, FLJ10639, FLJ21066, FLJ22268, FLJ43256, KIAA07... 29.6 2.8
mmu:66407 Mrps15, 1500003E24Rik, 2410002B11Rik, Mprs15; mitoch... 29.3 3.8
tgo:TGME49_111030 hypothetical protein 29.3 3.8
tgo:TGME49_045670 pyruvate dehydrogenase, putative (EC:1.2.4.1) 28.9 4.8
hsa:51191 HERC5, CEB1, CEBP1; hect domain and RLD 5 28.5
dre:568032 rbp2a, CRBP, CRBPII, MGC173957, MGC77921, rbp2, wu:... 28.5 6.3
ath:AT3G04030 myb family transcription factor 27.7 9.8
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 77.8 bits (190), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 66/110 (60%), Gaps = 9/110 (8%)
Query 1 GFEAYTEGQLSKLMEHVAKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAA 60
F AYT QL ++M VA + DP VE +RF+RIKQ+ ++ + D A+ +A+EHA+ AA+
Sbjct 613 AFAAYTPKQLRQVMAVVASKIQDP-QVEQDRFDRIKQRMIEELEDSASQVAYEHAIAAAS 671
Query 61 ILTRNDAFSRKDLLNALQQTNYDDSFAIAEHLQEETVDFQNCGVTHSLAF 110
+L RNDA SRKDLL L+ + + L E F++ H+ AF
Sbjct 672 VLLRNDANSRKDLLRLLKSS--------STSLNETLKTFRDLKAVHADAF 713
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 22/45 (48%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 87 AIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQLSKLM 131
A+AE L EETVD + CG++HS+ G G + F AYT QL ++M
Sbjct 582 ALAEQLDEETVDLKQCGISHSVGVSGDGLLLAFAAYTPKQLRQVM 626
> ath:AT5G18240 MYR1; MYR1 (MYb-related protein 1); transcription
factor
Length=402
Score = 31.6 bits (70), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 1/50 (2%)
Query 1 GFEAYTEGQLSKLMEHVAKLLSDPSMVEPERFERIKQKQMKLVADPATSM 50
G EA T+ QLS+L+ V+ D S +EP+ + + +QM+ P +S+
Sbjct 202 GIEA-TKAQLSELVSKVSADYPDSSFLEPKELQNLHHQQMQKTYPPNSSL 250
> hsa:7022 TFAP2C, AP2-GAMMA, ERF1, TFAP2G, hAP-2g; transcription
factor AP-2 gamma (activating enhancer binding protein 2
gamma); K09177 transcription factor AP-2 gamma
Length=450
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 25/47 (53%), Gaps = 2/47 (4%)
Query 46 PATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFAIAEHL 92
PA + LEA A+ R DA+ R DLL L + D+ +AE+L
Sbjct 121 PAGLLPHLSGLEAGAVSARRDAYRRSDLL--LPHAHALDAAGLAENL 165
> mmu:17909 Myo10, AW048724, D15Ertd600e, mKIAA0799; myosin X;
K12559 myosin X
Length=2062
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Query 51 AFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFAIAEHL-QEETVDFQNCGVTH 106
A E + IL +N R DLLN L+++ +D + + EH+ D CG H
Sbjct 550 AGEVQYDVRGILEKNRDTFRDDLLNLLRESRFDFIYDLFEHVSSRNNQDTLKCGSKH 606
> hsa:4651 MYO10, FLJ10639, FLJ21066, FLJ22268, FLJ43256, KIAA0799,
MGC131988; myosin X; K12559 myosin X
Length=2058
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Query 51 AFEHALEAAAILTRNDAFSRKDLLNALQQTNYDDSFAIAEHL-QEETVDFQNCGVTH 106
A E + IL +N R DLLN L+++ +D + + EH+ D CG H
Sbjct 550 AGEVQYDVRGILEKNRDTFRDDLLNLLRESRFDFIYDLFEHVSSRNNQDTLKCGSKH 606
> mmu:66407 Mrps15, 1500003E24Rik, 2410002B11Rik, Mprs15; mitochondrial
ribosomal protein S15
Length=258
Score = 29.3 bits (64), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 30/72 (41%), Gaps = 22/72 (30%)
Query 34 RIKQKQM--KLVADPATSMAFEHALEAAAILTRN--------------------DAFSRK 71
+IKQ+Q+ K+V +P S E + A + RN RK
Sbjct 106 KIKQEQLMNKIVENPEDSRTLEAQIIALTVRIRNYEEHMQKHRKDKAHKRHLLMSIDRRK 165
Query 72 DLLNALQQTNYD 83
LL L+QTNYD
Sbjct 166 KLLKILRQTNYD 177
> tgo:TGME49_111030 hypothetical protein
Length=3738
Score = 29.3 bits (64), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 4/34 (11%)
Query 84 DSFAIAEHLQEETVDFQNCGVTHSLAFKGTGFHM 117
DSFA A H+Q + DF V SL F+G H+
Sbjct 2665 DSFARAPHMQTSSCDF----VFFSLLFRGLASHL 2694
> tgo:TGME49_045670 pyruvate dehydrogenase, putative (EC:1.2.4.1)
Length=635
Score = 28.9 bits (63), Expect = 4.8, Method: Composition-based stats.
Identities = 25/96 (26%), Positives = 41/96 (42%), Gaps = 8/96 (8%)
Query 8 GQLSKLMEHVAKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAAILTRNDA 67
G+ L+E + S+ +P+ +KQK+ +V DP S FE L+ +
Sbjct 487 GEGPTLIEALTYRFRGHSVADPDEMRAVKQKEAWVVRDPIKS--FEEELKRLGYASDETI 544
Query 68 FSRKDLLNALQQTNYDDSFAIAEHLQEETVDFQNCG 103
+ + + A+ DD+ AE E D Q CG
Sbjct 545 AATRAKVKAV----VDDAVKFAETSPEP--DVQECG 574
> hsa:51191 HERC5, CEB1, CEBP1; hect domain and RLD 5
Length=1024
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 22/52 (42%), Gaps = 0/52 (0%)
Query 76 ALQQTNYDDSFAIAEHLQEETVDFQNCGVTHSLAFKGTGFHMGFEAYTEGQL 127
L T D ++ E L + V+F CG +HS G F A GQL
Sbjct 224 GLGHTESKDDPSLIEGLDNQKVEFVACGGSHSALLTQDGLLFTFGAGKHGQL 275
> dre:568032 rbp2a, CRBP, CRBPII, MGC173957, MGC77921, rbp2, wu:fb69e02;
retinol binding protein 2a, cellular; K14622 retinol
binding protein 2
Length=135
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 46/118 (38%), Gaps = 17/118 (14%)
Query 26 MVEPERFERIKQKQMK-LVADPATSMAFEHALEAAAILTRNDAFSRKDLLNALQQTNYDD 84
M+ + FE + MK L D AT H + I+ D F K L NY+
Sbjct 11 MLSNDNFEDV----MKALDIDFATRKIAVHLKQTKVIVQNGDKFETKTLSTF---RNYEV 63
Query 85 SFAIAEHLQEETVDFQNCGVTHSLAF---------KGTGFHMGFEAYTEGQLSKLMEH 133
+F I E E+T N V + + KG + G++ + EG L L H
Sbjct 64 NFVIGEEFDEQTKGLDNRTVKTLVKWDGDKLVCVQKGEKENRGWKQWIEGDLLHLEIH 121
> ath:AT3G04030 myb family transcription factor
Length=394
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 53/115 (46%), Gaps = 6/115 (5%)
Query 1 GFEAYTEGQLSKLMEHVAKLLSDPSMVEPERFERIKQKQMKLVADPATSMAFEHALEAAA 60
G EA + QLS+L+ V+ + S +EP+ + + +QM+ P S+ E L ++
Sbjct 202 GIEA-AKVQLSELVSKVSAEYPNSSFLEPKELQNLCSQQMQTNYPPDCSL--ESCLTSSE 258
Query 61 ILTRNDAFSRKDLLNALQQTNYDDSFAIAEHLQEETVDFQNCGVTHSLAFKGTGF 115
+N + L +T DS + + + EE + FQ +T + +G +
Sbjct 259 GTQKNSKMLENNRLGL--RTYIGDSTSEQKEIMEEPL-FQRMELTWTEGLRGNPY 310
Lambda K H
0.317 0.130 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40