bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2755_orf1
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
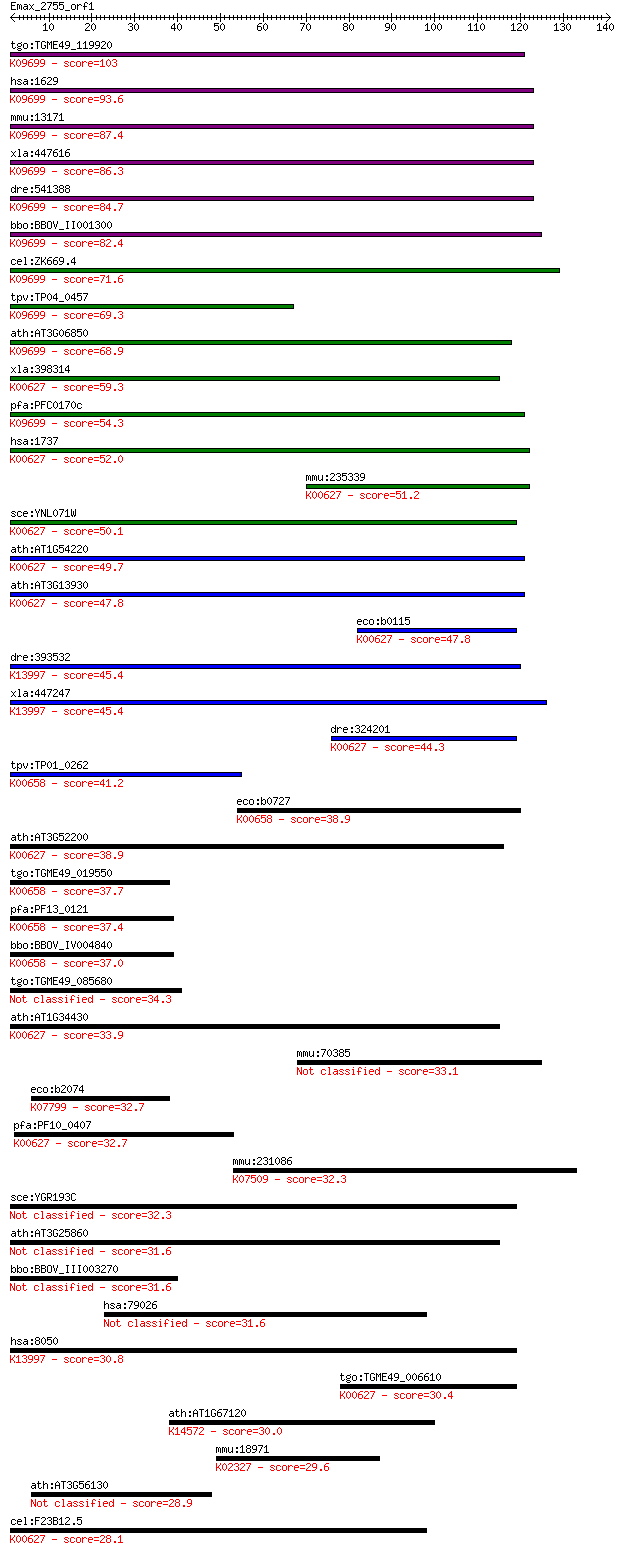
tgo:TGME49_119920 dihydrolipoamide branched chain transacylase... 103 2e-22
hsa:1629 DBT, BCATE2, E2, E2B, MGC9061; dihydrolipoamide branc... 93.6 2e-19
mmu:13171 Dbt, D3Wsu60e; dihydrolipoamide branched chain trans... 87.4 1e-17
xla:447616 dbt, MGC85493; dihydrolipoamide branched chain tran... 86.3 3e-17
dre:541388 dbt, im:7147214, zgc:103768; dihydrolipoamide branc... 84.7 7e-17
bbo:BBOV_II001300 18.m06098; lipoamide acyltransferase compone... 82.4 3e-16
cel:ZK669.4 hypothetical protein; K09699 2-oxoisovalerate dehy... 71.6 7e-13
tpv:TP04_0457 lipoamide transferase (EC:2.3.1.-); K09699 2-oxo... 69.3 3e-12
ath:AT3G06850 BCE2; BCE2; acetyltransferase/ alpha-ketoacid de... 68.9 5e-12
xla:398314 dlat; dihydrolipoamide S-acetyltransferase (EC:2.3.... 59.3 3e-09
pfa:PFC0170c dihydrolipoamide acyltransferase, putative (EC:2.... 54.3 1e-07
hsa:1737 DLAT, DLTA, PDC-E2, PDCE2; dihydrolipoamide S-acetylt... 52.0 5e-07
mmu:235339 Dlat, 6332404G05Rik, DLTA, PDC-E2; dihydrolipoamide... 51.2 9e-07
sce:YNL071W LAT1, ODP2, PDA2; Dihydrolipoamide acetyltransfera... 50.1 2e-06
ath:AT1G54220 dihydrolipoamide S-acetyltransferase, putative (... 49.7 3e-06
ath:AT3G13930 dihydrolipoamide S-acetyltransferase, putative (... 47.8 1e-05
eco:b0115 aceF, ECK0114, JW0111; pyruvate dehydrogenase, dihyd... 47.8 1e-05
dre:393532 pdhx, MGC66110, zgc:66110, zgc:85977; pyruvate dehy... 45.4 5e-05
xla:447247 pdhx, MGC86218; pyruvate dehydrogenase complex, com... 45.4 5e-05
dre:324201 dlat, wu:fc14f10, wu:fc21f08, wu:fc86g11, wu:fj57d0... 44.3 1e-04
tpv:TP01_0262 dihydrolipoamide succinyltransferase; K00658 2-o... 41.2 0.001
eco:b0727 sucB, ECK0715, JW0716; dihydrolipoyltranssuccinase (... 38.9 0.005
ath:AT3G52200 LTA3; LTA3; ATP binding / dihydrolipoyllysine-re... 38.9 0.005
tgo:TGME49_019550 dihydrolipoamide succinyltransferase compone... 37.7 0.012
pfa:PF13_0121 dihydrolipamide succinyltransferase component of... 37.4 0.014
bbo:BBOV_IV004840 23.m06243; dihydrolipoamide succinyltransfer... 37.0 0.020
tgo:TGME49_085680 dihydrolipoamide acyltransferase, putative (... 34.3 0.12
ath:AT1G34430 EMB3003 (embryo defective 3003); acyltransferase... 33.9 0.17
mmu:70385 Ccdc99, 1700018I02Rik, 2600001J17Rik, 2810049B11Rik,... 33.1 0.30
eco:b2074 mdtA, ECK2070, JW5338, yegM; multidrug efflux system... 32.7 0.37
pfa:PF10_0407 dihydrolipoamide acyltransferase, putative (EC:2... 32.7 0.38
mmu:231086 Hadhb, 4930479F15Rik, Mtpb; hydroxyacyl-Coenzyme A ... 32.3 0.50
sce:YGR193C PDX1; Dihydrolipoamide dehydrogenase (E3)-binding ... 32.3 0.52
ath:AT3G25860 LTA2; LTA2; dihydrolipoyllysine-residue acetyltr... 31.6 0.77
bbo:BBOV_III003270 17.m07312; biotin-requiring enzyme family p... 31.6 0.79
hsa:79026 AHNAK, AHNAKRS, MGC5395; AHNAK nucleoprotein 31.6 0.80
hsa:8050 PDHX, DLDBP, E3BP, OPDX, PDX1, proX; pyruvate dehydro... 30.8 1.5
tgo:TGME49_006610 biotin requiring domain-containing protein /... 30.4 1.7
ath:AT1G67120 ATP binding / ATPase/ nucleoside-triphosphatase/... 30.0 2.5
mmu:18971 Pold1, 125kDa; polymerase (DNA directed), delta 1, c... 29.6 3.3
ath:AT3G56130 biotin/lipoyl attachment domain-containing protein 28.9 5.5
cel:F23B12.5 hypothetical protein; K00627 pyruvate dehydrogena... 28.1 8.8
> tgo:TGME49_119920 dihydrolipoamide branched chain transacylase,
E2 subunit, putative (EC:2.3.1.168); K09699 2-oxoisovalerate
dehydrogenase E2 component (dihydrolipoyl transacylase)
[EC:2.3.1.168]
Length=510
Score = 103 bits (256), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 63/125 (50%), Positives = 79/125 (63%), Gaps = 11/125 (8%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEHA 60
SDKAAVEITSR++G IVKL+ +EG V++G PL+DID VE + P + A
Sbjct 110 SDKAAVEITSRFTGTIVKLHQKEGMMVRIGAPLMDID---VEAGEDHAEEEEPETKERPA 166
Query 61 AISQQQEQTTAAPSSKGGEAL-----ASPAVRRLAKEKGVDLDKVKGTGARGAITKDDIL 115
+S+ Q AA S G EA ASPA RR AKEKGVDL +VKG+G G ITK+D+L
Sbjct 167 PVSEPQ---AAASPSVGAEASSTTFSASPATRRFAKEKGVDLARVKGSGRNGLITKEDVL 223
Query 116 NYLSS 120
+L S
Sbjct 224 KFLES 228
> hsa:1629 DBT, BCATE2, E2, E2B, MGC9061; dihydrolipoamide branched
chain transacylase E2 (EC:2.3.1.168); K09699 2-oxoisovalerate
dehydrogenase E2 component (dihydrolipoyl transacylase)
[EC:2.3.1.168]
Length=482
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 55/122 (45%), Positives = 70/122 (57%), Gaps = 11/122 (9%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEHA 60
SDKA+V ITSRY G I KLY + VG PLVDI++ ++D++ P S EH
Sbjct 103 SDKASVTITSRYDGVIKKLYYNLDDIAYVGKPLVDIETEALKDSEEDVVETPAVSHDEHT 162
Query 61 AISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYLSS 120
QE KG + LA+PAVRRLA E + L +V G+G G I K+DILNYL
Sbjct 163 ----HQE-------IKGRKTLATPAVRRLAMENNIKLSEVVGSGKDGRILKEDILNYLEK 211
Query 121 ET 122
+T
Sbjct 212 QT 213
> mmu:13171 Dbt, D3Wsu60e; dihydrolipoamide branched chain transacylase
E2 (EC:2.3.1.168); K09699 2-oxoisovalerate dehydrogenase
E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=482
Score = 87.4 bits (215), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 51/122 (41%), Positives = 70/122 (57%), Gaps = 11/122 (9%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEHA 60
SDKA+V ITSRY G I +LY + VG PL+DI++ ++D++ P S EH
Sbjct 103 SDKASVTITSRYDGVIKRLYYNLDDIAYVGKPLIDIETEALKDSEEDVVETPAVSHDEHT 162
Query 61 AISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYLSS 120
QE KG + LA+PAVRRLA E + L +V G+G G I K+DIL++L
Sbjct 163 ----HQE-------IKGQKTLATPAVRRLAMENNIKLSEVVGSGKDGRILKEDILSFLEK 211
Query 121 ET 122
+T
Sbjct 212 QT 213
> xla:447616 dbt, MGC85493; dihydrolipoamide branched chain transacylase
E2 (EC:2.3.1.168); K09699 2-oxoisovalerate dehydrogenase
E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=492
Score = 86.3 bits (212), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 54/124 (43%), Positives = 71/124 (57%), Gaps = 13/124 (10%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSA--PPSSDSE 58
SDKA+V ITSRY G I KL+ ET VG PLVDI++ ++D ++ P S E
Sbjct 102 SDKASVTITSRYDGVIRKLHYNVDETAYVGKPLVDIETDALKDVAPEEDVVETPAVSHDE 161
Query 59 HAAISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYL 118
H QE KG + LA+PAVRRLA E + L +V G+G G I K+DIL +L
Sbjct 162 HT----HQE-------IKGHKTLATPAVRRLAMENNIKLSEVVGSGKDGRILKEDILGFL 210
Query 119 SSET 122
+ +T
Sbjct 211 AKQT 214
> dre:541388 dbt, im:7147214, zgc:103768; dihydrolipoamide branched
chain transacylase E2 (EC:2.3.1.168); K09699 2-oxoisovalerate
dehydrogenase E2 component (dihydrolipoyl transacylase)
[EC:2.3.1.168]
Length=493
Score = 84.7 bits (208), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 53/122 (43%), Positives = 70/122 (57%), Gaps = 9/122 (7%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEHA 60
SDKA+V ITSRY G I KLY VG PLVDI+ T Q+ +P E
Sbjct 102 SDKASVTITSRYDGVIRKLYYDVDSIALVGKPLVDIE------TDGGQAESPQEDVVETP 155
Query 61 AISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYLSS 120
A+SQ++ + KG + A+PAVRRLA E + L +V GTG G I K+DILN+++
Sbjct 156 AVSQEEH---SPQEIKGHKTQATPAVRRLAMENNIKLSEVVGTGKDGRILKEDILNFIAK 212
Query 121 ET 122
+T
Sbjct 213 QT 214
> bbo:BBOV_II001300 18.m06098; lipoamide acyltransferase component
of branched-chain alpha-keto acid dehydrogenase complex
(EC:2.3.1.12); K09699 2-oxoisovalerate dehydrogenase E2 component
(dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=417
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 52/129 (40%), Positives = 79/129 (61%), Gaps = 17/129 (13%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPD-----VEDTQAQQSSAPPSS 55
SDKAAV+ITSRY+G + KLY ++G+ +K+G PL+DID+ D E T+ +SS P
Sbjct 70 SDKAAVDITSRYTGLVKKLYVEQGKLIKIGSPLMDIDAEDDTPAVSEPTETTKSSIP--- 126
Query 56 DSEHAAISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDIL 115
S+ A S ++ S G A+P+VR+LAK+ GVD+ KV +G+ IT++D+
Sbjct 127 -SKPVAQSFKR--------SHGDSVRAAPSVRQLAKQLGVDITKVVPSGSNSQITREDVE 177
Query 116 NYLSSETST 124
+ +S S
Sbjct 178 KFAASSQSV 186
> cel:ZK669.4 hypothetical protein; K09699 2-oxoisovalerate dehydrogenase
E2 component (dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=448
Score = 71.6 bits (174), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 48/141 (34%), Positives = 73/141 (51%), Gaps = 29/141 (20%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSP-DVEDTQ------------AQ 47
SDKAAV I+ RY G + KLY + +VG L+D++ +VE+ + A
Sbjct 69 SDKAAVTISCRYDGIVKKLYHEVDGMARVGQALIDVEIEGNVEEPEQPKKEAASSSPEAP 128
Query 48 QSSAPPSSDSEHAAISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARG 107
+SSAP + +S H+ G+ LA+PAVRR+A E + L +V+GTG G
Sbjct 129 KSSAPKAPESAHSE----------------GKVLATPAVRRIAIENKIKLAEVRGTGKDG 172
Query 108 AITKDDILNYLSSETSTGTQG 128
+ K+D+L +L + T G
Sbjct 173 RVLKEDVLKFLGQVPADHTSG 193
> tpv:TP04_0457 lipoamide transferase (EC:2.3.1.-); K09699 2-oxoisovalerate
dehydrogenase E2 component (dihydrolipoyl transacylase)
[EC:2.3.1.168]
Length=420
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 36/68 (52%), Positives = 48/68 (70%), Gaps = 2/68 (2%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPD--VEDTQAQQSSAPPSSDSE 58
SDKAAVEITSRY+G + KLY +EGETVK+GGPL+DID+ D +DT SS S
Sbjct 79 SDKAAVEITSRYTGVVKKLYVKEGETVKIGGPLMDIDTVDEVPDDTPNNISSNLNDSKRH 138
Query 59 HAAISQQQ 66
++++ Q +
Sbjct 139 YSSVPQSK 146
> ath:AT3G06850 BCE2; BCE2; acetyltransferase/ alpha-ketoacid
dehydrogenase/ dihydrolipoamide branched chain acyltransferase
(EC:2.3.1.168); K09699 2-oxoisovalerate dehydrogenase E2
component (dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=483
Score = 68.9 bits (167), Expect = 5e-12, Method: Composition-based stats.
Identities = 43/121 (35%), Positives = 65/121 (53%), Gaps = 19/121 (15%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEHA 60
SDKA +EITSR+ GK+ + G+ +KVG LV + D +D+ S+
Sbjct 114 SDKATIEITSRFKGKVALISHSPGDIIKVGETLVRLAVEDSQDSLLTTDSS--------- 164
Query 61 AISQQQEQTTAAPSSKGGE----ALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILN 116
E T S +G E AL++PAVR LAK+ G+D++ + GTG G + K+D+L
Sbjct 165 ------EIVTLGGSKQGTENLLGALSTPAVRNLAKDLGIDINVITGTGKDGRVLKEDVLR 218
Query 117 Y 117
+
Sbjct 219 F 219
> xla:398314 dlat; dihydrolipoamide S-acetyltransferase (EC:2.3.1.12);
K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=628
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 43/132 (32%), Positives = 62/132 (46%), Gaps = 18/132 (13%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEG-ETVKVGGPLVDI--------------DSPDVEDTQ 45
+DKA + G + K+ EG V +G PL I +S V D +
Sbjct 234 TDKATIGFEVPEEGYLAKILVAEGTRDVPLGTPLCIIVEKESDISSFADYKESTGVVDIK 293
Query 46 AQQSSAPPSSDS---EHAAISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKG 102
Q + P++ S A+S +AAPS+ G SP ++LA EKG+D+ +VKG
Sbjct 294 PQHAPPTPTAASVPVPPVAVSTPAPTPSAAPSAPKGRVFVSPLAKKLAAEKGIDIKQVKG 353
Query 103 TGARGAITKDDI 114
+G G ITK DI
Sbjct 354 SGPEGRITKKDI 365
> pfa:PFC0170c dihydrolipoamide acyltransferase, putative (EC:2.3.1.-);
K09699 2-oxoisovalerate dehydrogenase E2 component
(dihydrolipoyl transacylase) [EC:2.3.1.168]
Length=448
Score = 54.3 bits (129), Expect = 1e-07, Method: Composition-based stats.
Identities = 38/120 (31%), Positives = 61/120 (50%), Gaps = 2/120 (1%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEHA 60
SDKAAV+ITS+Y+G +VK Y E + +KVG +ID+ D D + ++
Sbjct 72 SDKAAVDITSKYNGVLVKKYLNENDMLKVGSYFCEIDTDD--DIIERDEEEVEKEENNKK 129
Query 61 AISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYLSS 120
+ + + S ASP V+R AKE V+L+KV + I+ +D+ Y ++
Sbjct 130 EEDGESDLSLNDDISNNDYIKASPGVKRKAKEYKVNLNKVGDYFNKVNISLEDLELYYNN 189
> hsa:1737 DLAT, DLTA, PDC-E2, PDCE2; dihydrolipoamide S-acetyltransferase
(EC:2.3.1.12); K00627 pyruvate dehydrogenase E2
component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=647
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 60/140 (42%), Gaps = 19/140 (13%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEG-ETVKVGGPLV-------------DIDSPDVEDTQA 46
+DKA + + G + K+ EG V +G PL D +V D +
Sbjct 257 TDKATIGFEVQEEGYLAKILVPEGTRDVPLGTPLCIIVEKEADISAFADYRPTEVTDLKP 316
Query 47 QQSSAPPSSDSEHAAISQQQEQTTAAPSS-----KGGEALASPAVRRLAKEKGVDLDKVK 101
Q P + Q T +AP G SP ++LA EKG+DL +VK
Sbjct 317 QVPPPTPPPVAAVPPTPQPLAPTPSAPCPATPAGPKGRVFVSPLAKKLAVEKGIDLTQVK 376
Query 102 GTGARGAITKDDILNYLSSE 121
GTG G ITK DI +++ S+
Sbjct 377 GTGPDGRITKKDIDSFVPSK 396
> mmu:235339 Dlat, 6332404G05Rik, DLTA, PDC-E2; dihydrolipoamide
S-acetyltransferase (E2 component of pyruvate dehydrogenase
complex) (EC:2.3.1.12); K00627 pyruvate dehydrogenase E2
component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=642
Score = 51.2 bits (121), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 24/52 (46%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 70 TAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYLSSE 121
+AAP+ G SP ++LA EKG+DL +VKGTG G I K DI +++ S+
Sbjct 340 SAAPAGPKGRVFVSPLAKKLAAEKGIDLTQVKGTGPEGRIIKKDIDSFVPSK 391
> sce:YNL071W LAT1, ODP2, PDA2; Dihydrolipoamide acetyltransferase
component (E2) of pyruvate dehydrogenase complex, which
catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA
(EC:2.3.1.12); K00627 pyruvate dehydrogenase E2 component
(dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=482
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 64/140 (45%), Gaps = 22/140 (15%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEG-ETVKVGGPLV-----DIDSPDVEDTQAQQSSA--- 51
+DKA ++ + G + K+ EG + + V P+ D P +D + + S +
Sbjct 73 TDKAQMDFEFQEDGYLAKILVPEGTKDIPVNKPIAVYVEDKADVPAFKDFKLEDSGSDSK 132
Query 52 ------PPSSDSEHAAISQQQEQTTAAPSSKG-------GEALASPAVRRLAKEKGVDLD 98
P +E + +E T+AP +K G ASP + +A EKG+ L
Sbjct 133 TSTKAQPAEPQAEKKQEAPAEETKTSAPEAKKSDVAAPQGRIFASPLAKTIALEKGISLK 192
Query 99 KVKGTGARGAITKDDILNYL 118
V GTG RG ITK DI +YL
Sbjct 193 DVHGTGPRGRITKADIESYL 212
> ath:AT1G54220 dihydrolipoamide S-acetyltransferase, putative
(EC:2.3.1.12); K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=539
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 43/139 (30%), Positives = 61/139 (43%), Gaps = 20/139 (14%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVED----------------- 43
+DKA VE+ G + K+ EG G ++ I D ED
Sbjct 150 TDKATVEMECMEEGYLAKIVKAEGSKEIQVGEVIAITVEDEEDIGKFKDYTPSSTADAAP 209
Query 44 TQAQQSSAPPSSDSEHAAISQQQEQTTAAPSSK--GGEALASPAVRRLAKEKGVDLDKVK 101
T+A+ + APP + S E + PS+ G ASP R+LA++ V L ++
Sbjct 210 TKAEPTPAPPKEEKVKQP-SSPPEPKASKPSTPPTGDRVFASPLARKLAEDNNVPLSDIE 268
Query 102 GTGARGAITKDDILNYLSS 120
GTG G I K DI YL+S
Sbjct 269 GTGPEGRIVKADIDEYLAS 287
> ath:AT3G13930 dihydrolipoamide S-acetyltransferase, putative
(EC:2.3.1.12); K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=539
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 42/139 (30%), Positives = 66/139 (47%), Gaps = 20/139 (14%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEHA 60
+DKA VE+ G + K+ +EG G ++ I D +D Q + P SSD+ A
Sbjct 150 TDKATVEMECMEEGFLAKIVKEEGAKEIQVGEVIAITVEDEDDIQKFKDYTP-SSDTGPA 208
Query 61 AIS----------QQQEQTTAAPSSKGGE---------ALASPAVRRLAKEKGVDLDKVK 101
A ++ E+ +AP +K + ASP R+LA++ V L +K
Sbjct 209 APEAKPAPSLPKEEKVEKPASAPEAKISKPSSAPSEDRIFASPLARKLAEDNNVPLSSIK 268
Query 102 GTGARGAITKDDILNYLSS 120
GTG G I K D+ ++L+S
Sbjct 269 GTGPEGRIVKADVEDFLAS 287
> eco:b0115 aceF, ECK0114, JW0111; pyruvate dehydrogenase, dihydrolipoyltransacetylase
component E2 (EC:2.3.1.12); K00627
pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase)
[EC:2.3.1.12]
Length=630
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/37 (54%), Positives = 29/37 (78%), Gaps = 0/37 (0%)
Query 82 ASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYL 118
A+P +RRLA+E GV+L KVKGTG +G I ++D+ Y+
Sbjct 329 ATPLIRRLAREFGVNLAKVKGTGRKGRILREDVQAYV 365
> dre:393532 pdhx, MGC66110, zgc:66110, zgc:85977; pyruvate dehydrogenase
complex, component X; K13997 dihydrolipoamide dehydrogenase-binding
protein of pyruvate dehydrogenase complex
Length=490
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 40/134 (29%), Positives = 62/134 (46%), Gaps = 24/134 (17%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGET-VKVGGPL------------VDIDS--PDVEDTQ 45
+DKA V + S G + ++ QEG V++G + V+I + P T
Sbjct 101 TDKAVVVMESNEDGVLARILVQEGSRGVRLGTLIALMVSEGEDWKQVEIPALEPVTPPTA 160
Query 46 AQQSSAPPSSDSEHAAISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGA 105
A ++APP++ S A+ Q+ P + SPA R + G+D + +G
Sbjct 161 ALPTAAPPTAGSAPPAL----RQSVPTPLLR-----LSPAARHILDTHGLDPHQATASGP 211
Query 106 RGAITKDDILNYLS 119
RG ITK+D LN LS
Sbjct 212 RGIITKEDALNLLS 225
> xla:447247 pdhx, MGC86218; pyruvate dehydrogenase complex, component
X; K13997 dihydrolipoamide dehydrogenase-binding protein
of pyruvate dehydrogenase complex
Length=478
Score = 45.4 bits (106), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 36/126 (28%), Positives = 60/126 (47%), Gaps = 6/126 (4%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEG-ETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEH 59
+DKA V + S G + K+ +EG + V++G + + VE+ Q + PS
Sbjct 82 TDKAVVTMESNDDGVLAKILVEEGSKNVRLGSLIALL----VEEGQDWKQVHVPSVKVSP 137
Query 60 AAISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYLS 119
++ + AP +K G + SPA R + G+D + +G RG ITK+D L L+
Sbjct 138 TTVAAATKIANVAPVAKRGLRM-SPAARHIIDTHGLDTGSITPSGPRGIITKEDALKCLA 196
Query 120 SETSTG 125
+ G
Sbjct 197 QKEVPG 202
> dre:324201 dlat, wu:fc14f10, wu:fc21f08, wu:fc86g11, wu:fj57d06;
dihydrolipoamide S-acetyltransferase (E2 component of pyruvate
dehydrogenase complex) (EC:2.3.1.12); K00627 pyruvate
dehydrogenase E2 component (dihydrolipoamide acetyltransferase)
[EC:2.3.1.12]
Length=652
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/43 (46%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 76 KGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYL 118
+ G ASP ++LA EKGVD+ +V GTG G +TK DI +++
Sbjct 349 RKGRVFASPLAKKLAAEKGVDITQVTGTGPDGRVTKKDIDSFV 391
> tpv:TP01_0262 dihydrolipoamide succinyltransferase; K00658 2-oxoglutarate
dehydrogenase E2 component (dihydrolipoamide succinyltransferase)
[EC:2.3.1.61]
Length=456
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 34/57 (59%), Gaps = 3/57 (5%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDID---SPDVEDTQAQQSSAPPS 54
+DK +V++ S +SG + K ++ G+T+ VG PLV+ID P + + + PP+
Sbjct 111 TDKVSVDVNSPFSGVLTKTFSNTGDTILVGKPLVEIDLAGKPSDKPPEKKTEDKPPT 167
> eco:b0727 sucB, ECK0715, JW0716; dihydrolipoyltranssuccinase
(EC:2.3.1.61); K00658 2-oxoglutarate dehydrogenase E2 component
(dihydrolipoamide succinyltransferase) [EC:2.3.1.61]
Length=405
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 25/66 (37%), Positives = 39/66 (59%), Gaps = 2/66 (3%)
Query 54 SSDSEHAAISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDD 113
S+ SE A + Q Q A+ + +AL SPA+RRL E +D +KGTG G +T++D
Sbjct 88 SAKSEEKASTPAQRQQ-ASLEEQNNDAL-SPAIRRLLAEHNLDASAIKGTGVGGRLTRED 145
Query 114 ILNYLS 119
+ +L+
Sbjct 146 VEKHLA 151
> ath:AT3G52200 LTA3; LTA3; ATP binding / dihydrolipoyllysine-residue
acetyltransferase; K00627 pyruvate dehydrogenase E2
component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=637
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 58/117 (49%), Gaps = 5/117 (4%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEG-ETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEH 59
+DKA +E S G + K+ EG + V VG P+ I D E +A +SS+ SS+ +
Sbjct 251 TDKATLEFESLEEGYLAKILIPEGSKDVAVGKPIALI-VEDAESIEAIKSSSAGSSEVD- 308
Query 60 AAISQQQEQTTAAPSS-KGGEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDIL 115
+ + + P+ K G SPA + L E G++ ++ +G G + K D++
Sbjct 309 -TVKEVPDSVVDKPTERKAGFTKISPAAKLLILEHGLEASSIEASGPYGTLLKSDVV 364
> tgo:TGME49_019550 dihydrolipoamide succinyltransferase component
of 2-oxoglutaratedehydrogenase complex, putative (EC:2.3.1.61);
K00658 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide
succinyltransferase) [EC:2.3.1.61]
Length=470
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDID 37
+DK +V+I + +G+IV+ A G+TV+VG PL ID
Sbjct 132 TDKVSVDINAPQAGRIVRFEANAGDTVEVGKPLYVID 168
> pfa:PF13_0121 dihydrolipamide succinyltransferase component
of 2-oxoglutarate dehydrogenase complex (EC:2.3.1.61); K00658
2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide
succinyltransferase) [EC:2.3.1.61]
Length=421
Score = 37.4 bits (85), Expect = 0.014, Method: Composition-based stats.
Identities = 16/38 (42%), Positives = 26/38 (68%), Gaps = 0/38 (0%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDS 38
+DK +V+I S+ SG + K++A G+ V V PL +ID+
Sbjct 84 TDKVSVDINSKVSGGLSKIFADVGDVVLVDAPLCEIDT 121
> bbo:BBOV_IV004840 23.m06243; dihydrolipoamide succinyltransferase
(EC:2.3.1.61); K00658 2-oxoglutarate dehydrogenase E2
component (dihydrolipoamide succinyltransferase) [EC:2.3.1.61]
Length=402
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDS 38
+DK V+I S SG IVK + + +TV VG P +D+D+
Sbjct 94 TDKVTVDINSTLSGVIVKQHYEVDDTVLVGKPFIDVDA 131
> tgo:TGME49_085680 dihydrolipoamide acyltransferase, putative
(EC:2.4.1.115 2.3.1.61)
Length=470
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPD 40
+DK VEI S SG ++ AQEG+TV+VG L +D D
Sbjct 271 TDKVTVEIHSDCSGILLAQAAQEGDTVQVGSQLAVLDYSD 310
> ath:AT1G34430 EMB3003 (embryo defective 3003); acyltransferase/
dihydrolipoyllysine-residue acetyltransferase/ protein binding;
K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=465
Score = 33.9 bits (76), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 37/142 (26%), Positives = 62/142 (43%), Gaps = 30/142 (21%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSS---------- 50
SDKA +++ + Y G + + +EG VG + + + ED A +
Sbjct 78 SDKADMDVETFYDGYLAAIMVEEGGVAPVGSAIALL--AETEDEIADAKAKASGGGGGGD 135
Query 51 --------------APPSSDSEHAAISQQQEQTTAA---PSSKGGE-ALASPAVRRLAKE 92
AP S + + AA + A+ P+S+GG+ +ASP ++LAKE
Sbjct 136 SKAPPASPPTAAVEAPVSVEKKVAAAPVSIKAVAASAVHPASEGGKRIVASPYAKKLAKE 195
Query 93 KGVDLDKVKGTGARGAITKDDI 114
V+L + G+G G I D+
Sbjct 196 LKVELAGLVGSGPMGRIVAKDV 217
> mmu:70385 Ccdc99, 1700018I02Rik, 2600001J17Rik, 2810049B11Rik,
AA409762; coiled-coil domain containing 99
Length=608
Score = 33.1 bits (74), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 31/59 (52%), Gaps = 3/59 (5%)
Query 68 QTTAAPSSKGGEALASPAVRRLAKEKGVDL--DKVKGTGARGAITKDDILNYLSSETST 124
Q A P+SKG A RR+A E+ ++L DK + + A T+D + N + + ST
Sbjct 245 QQAADPNSKGNSLFAEVEDRRVAMERQLNLMKDKYQSLKKQNAFTRDQM-NKMKLQIST 302
> eco:b2074 mdtA, ECK2070, JW5338, yegM; multidrug efflux system,
subunit A; K07799 putative multidrug efflux transporter
MdtA
Length=415
Score = 32.7 bits (73), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 6 VEITSRYSGKIVKLYAQEGETVKVGGPLVDID 37
V + SR G+++ L+ QEG+ VK G L +ID
Sbjct 88 VTVRSRVDGQLIALHFQEGQQVKAGDLLAEID 119
> pfa:PF10_0407 dihydrolipoamide acyltransferase, putative (EC:2.3.1.12);
K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide
acetyltransferase) [EC:2.3.1.12]
Length=640
Score = 32.7 bits (73), Expect = 0.38, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 2 DKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQQSSAP 52
DK+ +E+ S YSG I KL +EG+ V + + I + +D + ++ P
Sbjct 222 DKSTIEVESPYSGIIKKLLVKEGQFVDLDKEVAIISITEEKDNEKEKIEEP 272
> mmu:231086 Hadhb, 4930479F15Rik, Mtpb; hydroxyacyl-Coenzyme
A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme
A hydratase (trifunctional protein), beta subunit (EC:2.3.1.16);
K07509 acetyl-CoA acyltransferase [EC:2.3.1.16]
Length=475
Score = 32.3 bits (72), Expect = 0.50, Method: Compositional matrix adjust.
Identities = 30/99 (30%), Positives = 44/99 (44%), Gaps = 19/99 (19%)
Query 53 PSSDSEHAAISQQQEQTTAA----------PSSKGGEALASPAVR--RLAKEKGVDLDKV 100
P+ A IS Q TTA + G E ++ +R R ++ +DL+K
Sbjct 131 PAHTVTMACISSNQAMTTAVGLIASGQCDVVVAGGVELMSDVPIRHSRNMRKMMLDLNKA 190
Query 101 KGTGARGAITKDDILNYLS------SETSTG-TQGHSAE 132
K G R ++ LN+LS +E ST T GHSA+
Sbjct 191 KTLGQRLSLLSKFRLNFLSPELPAVAEFSTNETMGHSAD 229
> sce:YGR193C PDX1; Dihydrolipoamide dehydrogenase (E3)-binding
protein (E3BP) of the mitochondrial pyruvate dehydrogenase
(PDH) complex, plays a structural role in the complex by binding
and positioning E3 to the dihydrolipoamide acetyltransferase
(E2) core
Length=410
Score = 32.3 bits (72), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 64/143 (44%), Gaps = 28/143 (19%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEG-ETVKVGGPLVDIDSPDVEDTQA-----QQSSA--- 51
+DK+ +++ + GK+ K+ EG + V VG P+ I DV+D A Q+++
Sbjct 71 TDKSQIDVEALDDGKLAKILKDEGSKDVDVGEPIAYI--ADVDDDLATIKLPQEANTANA 128
Query 52 ------PPSSDSEHAAISQQQEQTTAAP------SSKGGEALASPAVRRLAKEKGVD--- 96
PS+DS A Q ++ T P S E P+V L E +
Sbjct 129 KSIEIKKPSADSTEAT-QQHLKKATVTPIKTVDGSQANLEQTLLPSVSLLLAENNISKQK 187
Query 97 -LDKVKGTGARGAITKDDILNYL 118
L ++ +G+ G + K D+L YL
Sbjct 188 ALKEIAPSGSNGRLLKGDVLAYL 210
> ath:AT3G25860 LTA2; LTA2; dihydrolipoyllysine-residue acetyltransferase
Length=480
Score = 31.6 bits (70), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 33/127 (25%), Positives = 54/127 (42%), Gaps = 13/127 (10%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPL-------------VDIDSPDVEDTQAQ 47
SDKA +++ + Y G + + EGET VG + + + A+
Sbjct 94 SDKADMDVETFYDGYLAAIVVGEGETAPVGAAIGLLAETEAEIEEAKSKAASKSSSSVAE 153
Query 48 QSSAPPSSDSEHAAISQQQEQTTAAPSSKGGEALASPAVRRLAKEKGVDLDKVKGTGARG 107
P + A + Q A S + +A+P ++LAK+ VD++ V GTG G
Sbjct 154 AVVPSPPPVTSSPAPAIAQPAPVTAVSDGPRKTVATPYAKKLAKQHKVDIESVAGTGPFG 213
Query 108 AITKDDI 114
IT D+
Sbjct 214 RITASDV 220
> bbo:BBOV_III003270 17.m07312; biotin-requiring enzyme family
protein
Length=177
Score = 31.6 bits (70), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSP 39
+D+ V + S+ SG IV+ EG VKVG L+ I P
Sbjct 120 TDQVLVNVQSQLSGTIVETVGNEGCRVKVGADLIIIRRP 158
> hsa:79026 AHNAK, AHNAKRS, MGC5395; AHNAK nucleoprotein
Length=5890
Score = 31.6 bits (70), Expect = 0.80, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 35/76 (46%), Gaps = 1/76 (1%)
Query 23 EGETVKVGGPLVDIDSPDVEDTQAQQSSAPPSSDSEHAAISQQQ-EQTTAAPSSKGGEAL 81
+G V + G VDI++P++E T PS + IS + + AAP KGG +
Sbjct 530 KGPQVALKGSRVDIETPNLEGTLTGPRLGSPSGKTGTCRISMSEVDLNVAAPKVKGGVDV 589
Query 82 ASPAVRRLAKEKGVDL 97
P V K VD+
Sbjct 590 TLPRVEGKVKVPEVDV 605
> hsa:8050 PDHX, DLDBP, E3BP, OPDX, PDX1, proX; pyruvate dehydrogenase
complex, component X; K13997 dihydrolipoamide dehydrogenase-binding
protein of pyruvate dehydrogenase complex
Length=486
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 57/127 (44%), Gaps = 10/127 (7%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEG-ETVKVG---GPLVDIDSPDVEDTQAQQSSAPPSSD 56
+DKA V + + G + K+ +EG + +++G G +V+ + D + + + PP
Sbjct 80 TDKAVVTLDASDDGILAKIVVEEGSKNIRLGSLIGLIVE-EGEDWKHVEIPKDVGPPPPV 138
Query 57 SEHAAISQQQEQTTAAPSSK-----GGEALASPAVRRLAKEKGVDLDKVKGTGARGAITK 111
S+ + E + P K SPA R + ++ +D + TG RG TK
Sbjct 139 SKPSEPRPSPEPQISIPVKKEHIPGTLRFRLSPAARNILEKHSLDASQGTATGPRGIFTK 198
Query 112 DDILNYL 118
+D L +
Sbjct 199 EDALKLV 205
> tgo:TGME49_006610 biotin requiring domain-containing protein
/ 2-oxo acid dehydrogenases acyltransferase catalytic domain-containing
protein (EC:6.4.1.4 2.3.1.12 2.3.1.168); K00627
pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase)
[EC:2.3.1.12]
Length=932
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 78 GEALASPAVRRLAKEKGVDLDKVKGTGARGAITKDDILNYL 118
G+ LA+ LAK+ ++L++VKGTG IT D+ +L
Sbjct 610 GQPLATFNAIELAKKNKLNLEEVKGTGTNRRITAADVRQHL 650
> ath:AT1G67120 ATP binding / ATPase/ nucleoside-triphosphatase/
nucleotide binding / transcription factor binding; K14572
midasin
Length=5336
Score = 30.0 bits (66), Expect = 2.5, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 29/63 (46%), Gaps = 1/63 (1%)
Query 38 SPDVEDTQAQQSSAPPSSDSEHAAISQQQEQTTAAPSSKGGEALASPAVRRLAKEK-GVD 96
+ +VE A Q P S D +H + ++ T PS + EA A KE+ G D
Sbjct 4690 TEEVEKEDANQQEEPCSEDQKHPEEGENDQEETQEPSEENMEAEAEDRCGSPQKEEPGND 4749
Query 97 LDK 99
L++
Sbjct 4750 LEQ 4752
> mmu:18971 Pold1, 125kDa; polymerase (DNA directed), delta 1,
catalytic subunit (EC:2.7.7.7); K02327 DNA polymerase delta
subunit 1 [EC:2.7.7.7]
Length=1105
Score = 29.6 bits (65), Expect = 3.3, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 49 SSAPPSSDSEHAAISQQQEQTTAAPSSKGGEALASPAV 86
+ APP +EH + QQ+ + +GG+ L+ PAV
Sbjct 148 TPAPPGFGAEHLSELQQELNAAISRDQRGGKELSGPAV 185
> ath:AT3G56130 biotin/lipoyl attachment domain-containing protein
Length=194
Score = 28.9 bits (63), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 6 VEITSRYSGKIVKLYAQEGETVKVGGPLVDIDSPDVEDTQAQ 47
+ +TS +G+++KL + +G++V G PLV + P D Q
Sbjct 154 LPVTSDVAGEVLKLLSDDGDSVGYGDPLVAV-LPSFHDINIQ 194
> cel:F23B12.5 hypothetical protein; K00627 pyruvate dehydrogenase
E2 component (dihydrolipoamide acetyltransferase) [EC:2.3.1.12]
Length=507
Score = 28.1 bits (61), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 54/122 (44%), Gaps = 25/122 (20%)
Query 1 SDKAAVEITSRYSGKIVKLYAQEG-ETVKVGGPLVDI--DSPDV-------EDTQAQQSS 50
+DKA + + G + K+ QEG + V +G L I + DV +D + S
Sbjct 116 TDKATMGFETPEEGYLAKILIQEGSKDVPIGKLLCIIVDNEADVAAFKDFKDDGASSGGS 175
Query 51 AP----------PSSDSEHAAISQQQE-----QTTAAPSSKGGEALASPAVRRLAKEKGV 95
AP P++ S+ + +Q + ++ P S G ASP ++LA E G+
Sbjct 176 APAAEKAPEPAKPAASSQPSPPAQMYQAPSVPKSAPIPHSSSGRVSASPFAKKLAAENGL 235
Query 96 DL 97
DL
Sbjct 236 DL 237
Lambda K H
0.300 0.119 0.308
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2552834388
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40