bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2789_orf1
Length=152
Score E
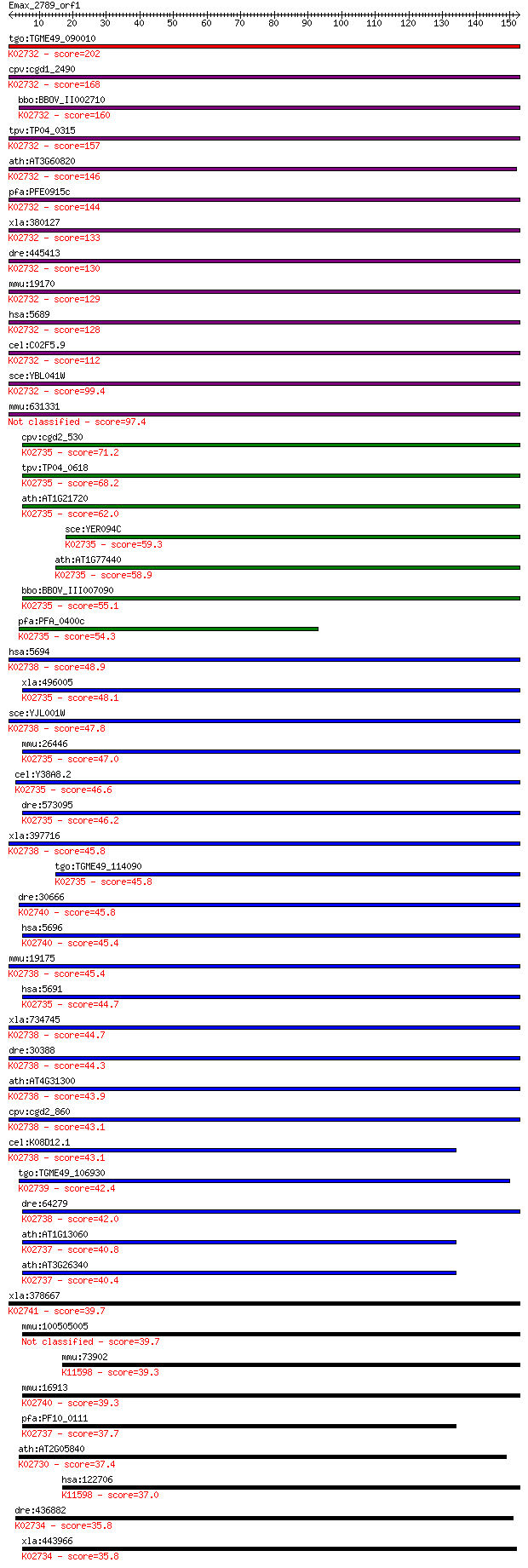
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_090010 proteasome subunit beta type 1, putative (EC... 202 3e-52
cpv:cgd1_2490 proteasome subunit beta type 1 ; K02732 20S prot... 168 6e-42
bbo:BBOV_II002710 18.m06221; proteasome A-type and B-type fami... 160 1e-39
tpv:TP04_0315 proteasome beat subunit; K02732 20S proteasome s... 157 1e-38
ath:AT3G60820 PBF1; PBF1; peptidase/ threonine-type endopeptid... 146 3e-35
pfa:PFE0915c proteasome subunit beta type 1, putative (EC:3.4.... 144 9e-35
xla:380127 psmb1, MGC154302, MGC52861; proteasome (prosome, ma... 133 3e-31
dre:445413 psmb1, MGC103665, wu:fc51f06, zgc:103665; proteasom... 130 1e-30
mmu:19170 Psmb1, AA409053, C81484, Lmpc5; proteasome (prosome,... 129 3e-30
hsa:5689 PSMB1, FLJ25321, HC5, KIAA1838, PMSB1, PSC5; proteaso... 128 7e-30
cel:C02F5.9 pbs-6; Proteasome Beta Subunit family member (pbs-... 112 5e-25
sce:YBL041W PRE7, PRS3; Beta 6 subunit of the 20S proteasome (... 99.4 4e-21
mmu:631331 Gm7061, EG631331; predicted gene 7061 97.4
cpv:cgd2_530 possible proteasome component ; K02735 20S protea... 71.2 1e-12
tpv:TP04_0618 proteasome subunit beta type 3 (EC:3.4.25.1); K0... 68.2 1e-11
ath:AT1G21720 PBC1; PBC1 (PROTEASOME BETA SUBUNIT C1); peptida... 62.0 7e-10
sce:YER094C PUP3, SCS32; Beta 3 subunit of the 20S proteasome ... 59.3 5e-09
ath:AT1G77440 PBC2; PBC2; peptidase/ threonine-type endopeptid... 58.9 6e-09
bbo:BBOV_III007090 17.m07625; proteasome A-type and B-type fam... 55.1 9e-08
pfa:PFA_0400c beta3 proteasome subunit, putative (EC:3.4.25.1)... 54.3 2e-07
hsa:5694 PSMB6, DELTA, LMPY, MGC5169, Y; proteasome (prosome, ... 48.9 5e-06
xla:496005 psmb3, HC10-II; proteasome (prosome, macropain) sub... 48.1 1e-05
sce:YJL001W PRE3, CRL21; Beta 1 subunit of the 20S proteasome,... 47.8 2e-05
mmu:26446 Psmb3, AL033320, C10-II, MGC106850; proteasome (pros... 47.0 3e-05
cel:Y38A8.2 pbs-3; Proteasome Beta Subunit family member (pbs-... 46.6 3e-05
dre:573095 psmb3, MGC56374, wu:fb11d10, wu:fu88b08, zgc:56374;... 46.2 5e-05
xla:397716 proteasome subunit Y; K02738 20S proteasome subunit... 45.8 5e-05
tgo:TGME49_114090 proteasome subunit beta type 3, putative (EC... 45.8 5e-05
dre:30666 psmb8, lmp7, macropain, prosome; proteasome (prosome... 45.8 6e-05
hsa:5696 PSMB8, D6S216, D6S216E, LMP7, MGC1491, PSMB5i, RING10... 45.4 6e-05
mmu:19175 Psmb6, Lmp19, Mpnd; proteasome (prosome, macropain) ... 45.4 8e-05
hsa:5691 PSMB3, HC10-II, MGC4147; proteasome (prosome, macropa... 44.7 1e-04
xla:734745 hypothetical protein MGC130855; K02738 20S proteaso... 44.7 1e-04
dre:30388 psmb6, y, zgc:109823; proteasome (prosome, macropain... 44.3 1e-04
ath:AT4G31300 PBA1; PBA1; endopeptidase/ peptidase/ threonine-... 43.9 2e-04
cpv:cgd2_860 Pre3p/proteasome regulatory subunit beta type 6, ... 43.1 3e-04
cel:K08D12.1 pbs-1; Proteasome Beta Subunit family member (pbs... 43.1 4e-04
tgo:TGME49_106930 proteasome subunit beta type 7, putative (EC... 42.4 5e-04
dre:64279 psmb11, MGC92422, macropain, prosome, zgc:92422; pro... 42.0 9e-04
ath:AT1G13060 PBE1; PBE1; endopeptidase/ peptidase/ threonine-... 40.8 0.002
ath:AT3G26340 20S proteasome beta subunit E, putative; K02737 ... 40.4 0.002
xla:378667 psmb9, LMP2, psmb9-A; proteasome (prosome, macropai... 39.7 0.003
mmu:100505005 proteasome subunit beta type-3-like 39.7 0.004
mmu:73902 Psmb11, 5830406J20Rik, beta5t; proteasome (prosome, ... 39.3 0.005
mmu:16913 Psmb8, Lmp-7, Lmp7; proteasome (prosome, macropain) ... 39.3 0.005
pfa:PF10_0111 20S proteasome beta subunit, putative; K02737 20... 37.7 0.015
ath:AT2G05840 PAA2; PAA2 (20S PROTEASOME SUBUNIT PAA2); endope... 37.4 0.017
hsa:122706 PSMB11, BETA5T, FLJ16369; proteasome (prosome, macr... 37.0 0.025
dre:436882 psmb2, zgc:92282; proteasome (prosome, macropain) s... 35.8 0.054
xla:443966 psmb2, MGC130720, MGC80364; proteasome (prosome, ma... 35.8 0.057
> tgo:TGME49_090010 proteasome subunit beta type 1, putative (EC:3.4.25.1);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=359
Score = 202 bits (514), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 94/152 (61%), Positives = 114/152 (75%), Gaps = 0/152 (0%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R SK+T+LT + +A+SGM+AD LH LK+RI Y HQHR PS+ +AQLLS VLY
Sbjct 185 RRQSKITKLTDKVVLATSGMEADKTTLHNLLKIRIEQYTHQHRHPPSLNAIAQLLSTVLY 244
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
SRRFFP+YTFNVL GID GKGAVYGYDA+GSFE S YN AGT LI +LDNQI+RNN
Sbjct 245 SRRFFPFYTFNVLCGIDENGKGAVYGYDAIGSFEPSRYNCAGTGAHLIMPVLDNQISRNN 304
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
Q E+ +T+E I++ + +ASAGERDIFTG
Sbjct 305 QQGEKAPLTRERIVEIVKSAVASAGERDIFTG 336
> cpv:cgd1_2490 proteasome subunit beta type 1 ; K02732 20S proteasome
subunit beta 6 [EC:3.4.25.1]
Length=246
Score = 168 bits (426), Expect = 6e-42, Method: Compositional matrix adjust.
Identities = 82/152 (53%), Positives = 105/152 (69%), Gaps = 1/152 (0%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R SK QLT +C +A SGM ADINAL K L ++ LY +H + PSI +AQLLSC+LY
Sbjct 73 RSVSKTCQLTNKCILACSGMLADINALRKTLLAKVKLYEFEHNKTPSINAIAQLLSCILY 132
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
S+RFFPYY+F +L G+D QGKG VYGYDAVGSF++ + + G+ SLITS+LDNQI+ NN
Sbjct 133 SKRFFPYYSFCLLSGLDEQGKGVVYGYDAVGSFDQHKFVALGSGGSLITSILDNQISGNN 192
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
Q + K II + + SA ERD+ TG
Sbjct 193 QT-SFESIDKVGIINIVKDSITSASERDVHTG 223
> bbo:BBOV_II002710 18.m06221; proteasome A-type and B-type family
protein; K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=217
Score = 160 bits (406), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 76/149 (51%), Positives = 102/149 (68%), Gaps = 0/149 (0%)
Query 4 SKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRR 63
SKL +LT C I +SGMQAD++ALH L+ +I LYR +H +P I +AQLLS +LYSRR
Sbjct 46 SKLKKLTQTCVIGTSGMQADMHALHSALERQIELYRFKHHREPGISVIAQLLSTILYSRR 105
Query 64 FFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLD 123
FFPYYTFN+L G+D G+G G+DA+G++ Y + GT++SLI S+LDN + NQ
Sbjct 106 FFPYYTFNILCGVDENGRGITCGFDAIGNYGFDAYMAKGTSSSLIMSILDNHLTSRNQQK 165
Query 124 ERPKMTKEEIIQFARGVLASAGERDIFTG 152
+ EE+I +G + SA ERDIFTG
Sbjct 166 HNEIQSLEELINVIKGAMTSAVERDIFTG 194
> tpv:TP04_0315 proteasome beat subunit; K02732 20S proteasome
subunit beta 6 [EC:3.4.25.1]
Length=218
Score = 157 bits (397), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 75/152 (49%), Positives = 103/152 (67%), Gaps = 0/152 (0%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R +KL +LT +C + +SGMQAD+ AL L+ +I LYR+ H +P+ +AQLLS VLY
Sbjct 44 RFSTKLFKLTDKCILGTSGMQADMLALQSVLERQIELYRYTHNREPTFNAIAQLLSTVLY 103
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
RRFFPYYTFN+L GID QGKG V GYDAVG++ Y + GT++SL+ S+LDN + NN
Sbjct 104 GRRFFPYYTFNILCGIDEQGKGVVCGYDAVGNYNYEKYTAQGTSSSLVISVLDNLLNGNN 163
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
Q + + E+++ + + SA ERDI TG
Sbjct 164 QKVKPEVNSLEDLVNLVKNAMVSAVERDICTG 195
> ath:AT3G60820 PBF1; PBF1; peptidase/ threonine-type endopeptidase;
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=223
Score = 146 bits (368), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 73/156 (46%), Positives = 100/156 (64%), Gaps = 5/156 (3%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R +SK+ +L R ++SSG QAD+ AL K LK R +Y+HQH + S +AQLLS LY
Sbjct 44 RDYSKIHKLADRAVLSSSGFQADVKALQKVLKSRHLIYQHQHNKQMSCPAMAQLLSNTLY 103
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
+RFFPYY FNVL G+D +GKG V+ YDAVGS+ER Y + G+ ++LI LDNQ+ +
Sbjct 104 FKRFFPYYAFNVLGGLDEEGKGCVFTYDAVGSYERVGYGAQGSGSTLIMPFLDNQLKSPS 163
Query 121 QL-----DERPKMTKEEIIQFARGVLASAGERDIFT 151
L D +++ E + + V ASA ERDI+T
Sbjct 164 PLLLPKQDSNTPLSEAEAVDLVKTVFASATERDIYT 199
> pfa:PFE0915c proteasome subunit beta type 1, putative (EC:3.4.25.1);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=240
Score = 144 bits (364), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 66/152 (43%), Positives = 97/152 (63%), Gaps = 0/152 (0%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R K+++LT +C I SSGMQ+DI LH L+ +I L+ +H P I +A+LL +LY
Sbjct 66 RFCPKISKLTDKCIIGSSGMQSDIKTLHSLLQKKIQLFVLEHSHYPDIHVIARLLCVILY 125
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
SRRFFPYY FN+L G+D KG +Y YD+VGS+ + ++ G+ + LI +LDN++ + N
Sbjct 126 SRRFFPYYAFNILAGVDENNKGVLYNYDSVGSYCEATHSCVGSGSQLILPILDNRVEQKN 185
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
QL + + I F + + SA ERDI+TG
Sbjct 186 QLIKNTNFNLGDDINFVKDAITSATERDIYTG 217
> xla:380127 psmb1, MGC154302, MGC52861; proteasome (prosome,
macropain) subunit, beta type, 1 (EC:3.4.25.1); K02732 20S proteasome
subunit beta 6 [EC:3.4.25.1]
Length=239
Score = 133 bits (334), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 62/153 (40%), Positives = 99/153 (64%), Gaps = 1/153 (0%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R K +LT + I +G AD L K ++ R+ +Y+H + + + +A +LS +LY
Sbjct 64 RNTPKCYKLTDKTVIGCTGFHADCLTLTKIIEARLKMYKHSNNKTMTSGAIAAMLSTILY 123
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIA-RN 119
SRRFFPYY +N++ G+D +GKGAVY +D VGS++R Y + G+A++++ LLDNQI +N
Sbjct 124 SRRFFPYYVYNIIGGLDEEGKGAVYSFDPVGSYQRDAYKAGGSASAMLQPLLDNQIGYKN 183
Query 120 NQLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
Q E+ +T E+ ++ + V SA ERD++TG
Sbjct 184 MQNVEQLPLTLEKALKLIKDVFISAAERDVYTG 216
> dre:445413 psmb1, MGC103665, wu:fc51f06, zgc:103665; proteasome
(prosome, macropain) subunit, beta type, 1 (EC:3.4.25.1);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=237
Score = 130 bits (328), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 59/153 (38%), Positives = 96/153 (62%), Gaps = 1/153 (0%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R K +LT + SG D L K ++ R+ +Y+H + + + +A +LS +LY
Sbjct 62 RDSPKCYKLTDTTVLGCSGFHGDCLTLTKIIEARLKMYKHSNNKSMTSGAIAAMLSTILY 121
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIA-RN 119
RRFFPYY +N++ G+D +G+GAVY +D VGS++R Y + G+A++++ LLDNQI +N
Sbjct 122 GRRFFPYYVYNIIGGLDEEGRGAVYSFDPVGSYQRDTYKAGGSASAMLQPLLDNQIGFKN 181
Query 120 NQLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
+ E +T+E+ +Q + V SA ERD++TG
Sbjct 182 MENVEHVPLTQEKAVQLVKDVFISAAERDVYTG 214
> mmu:19170 Psmb1, AA409053, C81484, Lmpc5; proteasome (prosome,
macropain) subunit, beta type 1 (EC:3.4.25.1); K02732 20S
proteasome subunit beta 6 [EC:3.4.25.1]
Length=240
Score = 129 bits (325), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 59/153 (38%), Positives = 97/153 (63%), Gaps = 1/153 (0%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R K +LT + I SG D L K ++ R+ +Y+H + + + +A +LS +LY
Sbjct 65 RDSPKCYKLTDKTVIGCSGFHGDCLTLTKIIEARLKMYKHSNNKAMTTGAIAAMLSTILY 124
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIA-RN 119
SRRFFPYY +N++ G+D +GKGAVY +D VGS++R ++ + G+A++++ LLDNQ+ +N
Sbjct 125 SRRFFPYYVYNIIGGLDEEGKGAVYSFDPVGSYQRDSFKAGGSASAMLQPLLDNQVGFKN 184
Query 120 NQLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
Q E +T + ++ + V SA ERD++TG
Sbjct 185 MQNVEHVPLTLDRAMRLVKDVFISAAERDVYTG 217
> hsa:5689 PSMB1, FLJ25321, HC5, KIAA1838, PMSB1, PSC5; proteasome
(prosome, macropain) subunit, beta type, 1 (EC:3.4.25.1);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=241
Score = 128 bits (321), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 58/153 (37%), Positives = 97/153 (63%), Gaps = 1/153 (0%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R K +LT + I SG D L K ++ R+ +Y+H + + + +A +LS +LY
Sbjct 66 RDSPKCYKLTDKTVIGCSGFHGDCLTLTKIIEARLKMYKHSNNKAMTTGAIAAMLSTILY 125
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIA-RN 119
SRRFFPYY +N++ G+D +GKGAVY +D VGS++R ++ + G+A++++ LLDNQ+ +N
Sbjct 126 SRRFFPYYVYNIIGGLDEEGKGAVYSFDPVGSYQRDSFKAGGSASAMLQPLLDNQVGFKN 185
Query 120 NQLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
Q E ++ + ++ + V SA ERD++TG
Sbjct 186 MQNVEHVPLSLDRAMRLVKDVFISAAERDVYTG 218
> cel:C02F5.9 pbs-6; Proteasome Beta Subunit family member (pbs-6);
K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=258
Score = 112 bits (280), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 62/154 (40%), Positives = 85/154 (55%), Gaps = 2/154 (1%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R K+ L + +SG D+ L K L+ R+ YR +R D S++ A+LLS LY
Sbjct 81 RDAEKIQILNDNIILTTSGFYGDVLQLKKVLQSRLHKYRFDYRSDMSVDLCAELLSRNLY 140
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
RRFFPYYT +L GID GKGAV+ YD +G ER Y+++G A +I LD QI
Sbjct 141 YRRFFPYYTGAILAGIDEHGKGAVFSYDPIGCIERLGYSASGAAEPMIIPFLDCQIGHVT 200
Query 121 QLD--ERPKMTKEEIIQFARGVLASAGERDIFTG 152
+ ERP++T + I + A ER+I TG
Sbjct 201 LSEGYERPELTLDRAISLMKDSFRGAAEREISTG 234
> sce:YBL041W PRE7, PRS3; Beta 6 subunit of the 20S proteasome
(EC:3.4.25.1); K02732 20S proteasome subunit beta 6 [EC:3.4.25.1]
Length=241
Score = 99.4 bits (246), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 58/162 (35%), Positives = 84/162 (51%), Gaps = 10/162 (6%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDP-SIEGLAQLLSCVL 59
R K+ ++++G AD +AL K K + Y H + SI A+ + +L
Sbjct 57 RYEPKVFDCGDNIVMSANGFAADGDALVKRFKNSVKWYHFDHNDKKLSINSAARNIQHLL 116
Query 60 YSRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARN 119
Y +RFFPYY ++ G+D GKGAVY +D VGS+ER + G A SLI LDNQ+
Sbjct 117 YGKRFFPYYVHTIIAGLDEDGKGAVYSFDPVGSYEREQCRAGGAAASLIMPFLDNQVNFK 176
Query 120 NQLD-------ERP--KMTKEEIIQFARGVLASAGERDIFTG 152
NQ + ++P ++ EE+I+ R SA ER I G
Sbjct 177 NQYEPGTNGKVKKPLKYLSVEEVIKLVRDSFTSATERHIQVG 218
> mmu:631331 Gm7061, EG631331; predicted gene 7061
Length=730
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 51/154 (33%), Positives = 85/154 (55%), Gaps = 8/154 (5%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R K +LT + I SG D L K ++ R+ + + +A +LS +
Sbjct 65 RDSPKCYKLTDKTVIGCSGFHGDCLTLTKIIETRLKMNKAMR-----TGAVAAMLSTIPC 119
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIA--R 118
SRRFFPYY +N++ G+D +GKGAVY +D VGS++R + + +A++ + LL NQ R
Sbjct 120 SRRFFPYYVYNIIGGLDEEGKGAVYSFDPVGSYQRDSVKAGASASATLQPLLHNQTGFRR 179
Query 119 NNQLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
++ P++ + ++ + V SA ERD++TG
Sbjct 180 MQSVEHVPRIL-DRAMRLVKDVFISAAERDVYTG 212
> cpv:cgd2_530 possible proteasome component ; K02735 20S proteasome
subunit beta 3 [EC:3.4.25.1]
Length=204
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 76/149 (51%), Gaps = 9/149 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ + + + + SG+ DI+ L +K + +LY + + ++ L+ + + +LYS+RF
Sbjct 41 KVIRPSSKTLMGFSGLATDIHTLTNLIKFKTNLYHLREEREIGVKALSHMTASILYSKRF 100
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGSFER-SNYNSAGTANSLITSLLDNQIARNNQLD 123
PY+ ++ G+DN + GYD +G S + +GTA DNQ+ +
Sbjct 101 SPYFVEPIVAGLDNNNVPFIAGYDLIGCLSVCSEFAISGTA--------DNQLFGICESY 152
Query 124 ERPKMTKEEIIQFARGVLASAGERDIFTG 152
RP + +E+++ + S +RD F+G
Sbjct 153 YRPDLEPDELMEIVSQCMLSGIDRDAFSG 181
> tpv:TP04_0618 proteasome subunit beta type 3 (EC:3.4.25.1);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=206
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/149 (24%), Positives = 78/149 (52%), Gaps = 9/149 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K ++T C A+SG+ D+ L + + ++++Y+ + ++ S++ L+ ++ +LYSRRF
Sbjct 43 KAFKVTESCFFAASGLATDVQTLKEEIMFKVNMYKLRGDKEMSVKTLSNMVGAMLYSRRF 102
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGS-FERSNYNSAGTANSLITSLLDNQIARNNQLD 123
P++ +V+ G+DN + +D VG+ S++ AGT + + + +
Sbjct 103 GPWFVDSVIAGLDNDSSPYITCFDLVGAPCTPSDFVVAGTCSEQLYGVCEALF------- 155
Query 124 ERPKMTKEEIIQFARGVLASAGERDIFTG 152
+P M E++ + L + +RD +G
Sbjct 156 -KPGMDPEQLFETVSQCLMAGIDRDCLSG 183
> ath:AT1G21720 PBC1; PBC1 (PROTEASOME BETA SUBUNIT C1); peptidase/
threonine-type endopeptidase; K02735 20S proteasome subunit
beta 3 [EC:3.4.25.1]
Length=204
Score = 62.0 bits (149), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 73/149 (48%), Gaps = 9/149 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
+++++ R I SG+ D+ L++ L R LY+ + D E A L+S +LY +RF
Sbjct 41 RISKIHDRVFIGLSGLATDVQTLYQRLVFRHKLYQLREERDMKPETFASLVSAILYEKRF 100
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGSFERS-NYNSAGTANSLITSLLDNQIARNNQLD 123
PY V+ G+ + K + D++G+ E + ++ +GTA+ + +
Sbjct 101 GPYLCQPVIAGLGDDDKPFICTMDSIGAKELAKDFVVSGTASESLYGACEAMY------- 153
Query 124 ERPKMTKEEIIQFARGVLASAGERDIFTG 152
+P M EE+ + L S+ +RD +G
Sbjct 154 -KPDMEAEELFETISQALLSSVDRDCLSG 181
> sce:YER094C PUP3, SCS32; Beta 3 subunit of the 20S proteasome
involved in ubiquitin-dependent catabolism; human homolog
is subunit C10 (EC:3.4.25.1); K02735 20S proteasome subunit
beta 3 [EC:3.4.25.1]
Length=205
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 69/137 (50%), Gaps = 10/137 (7%)
Query 18 SGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRFFPYYTFNVLFGID 77
+G+ D+ L++ + + +LY+ + E QL+S LY RRF PY+ V+ GI+
Sbjct 54 TGLATDVTTLNEMFRYKTNLYKLKEERAIEPETFTQLVSSSLYERRFGPYFVGPVVAGIN 113
Query 78 NQ-GKGAVYGYDAVGSF-ERSNYNSAGTANSLITSLLDNQIARNNQLDERPKMTKEEIIQ 135
++ GK + G+D +G E ++ +GTA+ + + ++ P + E++ +
Sbjct 114 SKSGKPFIAGFDLIGCIDEAKDFIVSGTASDQLFGMCESLY--------EPNLEPEDLFE 165
Query 136 FARGVLASAGERDIFTG 152
L +A +RD +G
Sbjct 166 TISQALLNAADRDALSG 182
> ath:AT1G77440 PBC2; PBC2; peptidase/ threonine-type endopeptidase;
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=204
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 37/139 (26%), Positives = 67/139 (48%), Gaps = 9/139 (6%)
Query 15 IASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRFFPYYTFNVLF 74
I SG+ D+ L++ L R LY+ + D E A L+S +LY +RF P+ V+
Sbjct 51 IGLSGLATDVQTLYQRLVFRHKLYQLREERDMKPETFASLVSAILYEKRFGPFLCQPVIA 110
Query 75 GIDNQGKGAVYGYDAVGSFERS-NYNSAGTANSLITSLLDNQIARNNQLDERPKMTKEEI 133
G+ + K + D++G+ E + ++ +GTA+ + + +P M EE+
Sbjct 111 GLGDDNKPFICTMDSIGAKELAKDFVVSGTASESLYGACEAMF--------KPDMEAEEL 162
Query 134 IQFARGVLASAGERDIFTG 152
+ L S+ +RD +G
Sbjct 163 FETISQALLSSVDRDCLSG 181
> bbo:BBOV_III007090 17.m07625; proteasome A-type and B-type family
protein; K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 55.1 bits (131), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 33/149 (22%), Positives = 72/149 (48%), Gaps = 10/149 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K ++T A+ G+ DI + ++ + ++Y + ++ ++ LA ++ +LYSRRF
Sbjct 43 KAFKVTDTAYFAACGLATDIQTMKSEIEFKTNMYALRCEKNMGVKTLAHMVGSLLYSRRF 102
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGS-FERSNYNSAGTANSLITSLLDNQIARNNQLD 123
P++ +V+ G+D +Y +D +G+ ++ GT + + + ++
Sbjct 103 GPWFVSSVVAGLDKDTP-HIYCFDLIGAPCNAKDFVVVGTCSEQLYGICESLY------- 154
Query 124 ERPKMTKEEIIQFARGVLASAGERDIFTG 152
RP M E+ + L +A +RD +G
Sbjct 155 -RPNMEPSELFETISQCLMAAIDRDCLSG 182
> pfa:PFA_0400c beta3 proteasome subunit, putative (EC:3.4.25.1);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=204
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 52/101 (51%), Gaps = 12/101 (11%)
Query 4 SKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRR 63
SK+ ++ + SG+ DI L++ L+ R++LY + + +E A +LS +LYS R
Sbjct 28 SKIFKMNNNVYVGLSGLATDIQTLYEILRYRVNLYEVRQDAEMDVECFANMLSSILYSNR 87
Query 64 FFPYYTFNVLFG------IDNQGKGAV------YGYDAVGS 92
F PY+ ++ G +D +G+ V YD +G+
Sbjct 88 FSPYFVNPIVVGFKLKHYVDEEGEKKVNYEPYLTAYDLIGA 128
> hsa:5694 PSMB6, DELTA, LMPY, MGC5169, Y; proteasome (prosome,
macropain) subunit, beta type, 6 (EC:3.4.25.1); K02738 20S
proteasome subunit beta 1 [EC:3.4.25.1]
Length=239
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 43/152 (28%), Positives = 62/152 (40%), Gaps = 9/152 (5%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R+ KLT + R SG AD A+ + ++ + + E P + A L + Y
Sbjct 63 RVTDKLTPIHDRIFCCRSGSAADTQAVADAVTYQLGFHSIELNEPPLVHTAASLFKEMCY 122
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
R ++ G D Q G VY G R ++ G+ +S I +D
Sbjct 123 RYRE-DLMAGIIIAGWDPQEGGQVYSVPMGGMMVRQSFAIGGSGSSYIYGYVDATY---- 177
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
R MTKEE +QF LA A ERD +G
Sbjct 178 ----REGMTKEECLQFTANALALAMERDGSSG 205
> xla:496005 psmb3, HC10-II; proteasome (prosome, macropain) subunit,
beta type, 3; K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 33/150 (22%), Positives = 69/150 (46%), Gaps = 10/150 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ + R I +G+ D+ + + LK R++LY + + +++ +LY RRF
Sbjct 41 KIFPMGERLYIGLAGLATDVQTVAQRLKFRLNLYELKEGRQIKPKTFMSMVANLLYERRF 100
Query 65 FPYYTFNVLFGIDNQG-KGAVYGYDAVGS-FERSNYNSAGTANSLITSLLDNQIARNNQL 122
PYY V+ G+D + + + D +G E ++ +GT + + + ++
Sbjct 101 GPYYIEPVIAGLDPKTFQPFICSLDLIGCPMETEDFVVSGTCSEQMYGMCESLW------ 154
Query 123 DERPKMTKEEIIQFARGVLASAGERDIFTG 152
P M E++ + + +A +RD +G
Sbjct 155 --EPDMEPEDLFETISQAMLNAVDRDAVSG 182
> sce:YJL001W PRE3, CRL21; Beta 1 subunit of the 20S proteasome,
responsible for cleavage after acidic residues in peptides
(EC:3.4.25.1); K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=215
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 41/153 (26%), Positives = 67/153 (43%), Gaps = 12/153 (7%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R+ KLT++ + SG AD A+ ++ + LY Q+ PS E A + + Y
Sbjct 48 RVTDKLTRVHDKIWCCRSGSAADTQAIADIVQYHLELYTSQYGT-PSTETAASVFKELCY 106
Query 61 SRRFFPYYTFNVLF-GIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARN 119
+ T ++ G D++ KG VY GS + Y AG+ ++ I D
Sbjct 107 ENK--DNLTAGIIVAGYDDKNKGEVYTIPLGGSVHKLPYAIAGSGSTFIYGYCDKNF--- 161
Query 120 NQLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
R M+KEE + F + L+ A + D +G
Sbjct 162 -----RENMSKEETVDFIKHSLSQAIKWDGSSG 189
> mmu:26446 Psmb3, AL033320, C10-II, MGC106850; proteasome (prosome,
macropain) subunit, beta type 3 (EC:3.4.25.1); K02735
20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 34/150 (22%), Positives = 68/150 (45%), Gaps = 10/150 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ + R I +G+ D+ + + LK R++LY + L +++ +LY +RF
Sbjct 41 KIFPMGDRLYIGLAGLATDVQTVAQRLKFRLNLYELKEGRQIKPYTLMSMVANLLYEKRF 100
Query 65 FPYYTFNVLFGIDNQG-KGAVYGYDAVGS-FERSNYNSAGTANSLITSLLDNQIARNNQL 122
PYYT V+ G+D + K + D +G ++ +GT + + + ++
Sbjct 101 GPYYTEPVIAGLDPKTFKPFICSLDLIGCPMVTDDFVVSGTCSEQMYGMCESL------- 153
Query 123 DERPKMTKEEIIQFARGVLASAGERDIFTG 152
P M E + + + +A +RD +G
Sbjct 154 -WEPNMDPEHLFETISQAMLNAVDRDAVSG 182
> cel:Y38A8.2 pbs-3; Proteasome Beta Subunit family member (pbs-3);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=204
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 33/151 (21%), Positives = 70/151 (46%), Gaps = 9/151 (5%)
Query 3 HSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSR 62
K+ ++T + + +G Q+D + + + R +LY + + + L++++S + Y
Sbjct 39 QKKVHKVTDKVYVGLAGFQSDARTVLEKIMFRKNLYELRENRNIKPQVLSEMISNLAYQH 98
Query 63 RFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERS-NYNSAGTANSLITSLLDNQIARNNQ 121
RF Y+T ++ G+D+ K + D +G ++ + GT + + +N N +
Sbjct 99 RFGSYFTEPLVAGLDDTNKPYICCMDTIGCVSAPRDFVAVGTGQEYLLGVCENFWRENMK 158
Query 122 LDERPKMTKEEIIQFARGVLASAGERDIFTG 152
DE + T + I+ S ERD +G
Sbjct 159 PDELFEATAQSIL--------SCLERDAASG 181
> dre:573095 psmb3, MGC56374, wu:fb11d10, wu:fu88b08, zgc:56374;
proteasome (prosome, macropain) subunit, beta type, 3 (EC:3.4.25.1);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 33/150 (22%), Positives = 67/150 (44%), Gaps = 10/150 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ + R I +G+ D+ + + LK R++LY + ++S +LY RRF
Sbjct 41 KIFPMGERLYIGLAGLATDVQTVSQRLKFRLNLYELKEGRQIKPRTFMSMVSNLLYERRF 100
Query 65 FPYYTFNVLFGIDNQG-KGAVYGYDAVGS-FERSNYNSAGTANSLITSLLDNQIARNNQL 122
PYY V+ G+D + + + D +G ++ +GT + + + ++
Sbjct 101 GPYYIEPVIAGLDPKTFEPFICSLDLIGCPMVTEDFVVSGTCSEQMYGMCESLW------ 154
Query 123 DERPKMTKEEIIQFARGVLASAGERDIFTG 152
P M E++ + + +A +RD +G
Sbjct 155 --EPDMKPEDLFETISQAMLNAVDRDAVSG 182
> xla:397716 proteasome subunit Y; K02738 20S proteasome subunit
beta 1 [EC:3.4.25.1]
Length=233
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 40/152 (26%), Positives = 62/152 (40%), Gaps = 9/152 (5%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R+ KLT + R SG AD A+ + ++ + + P + A L + Y
Sbjct 62 RVTDKLTPVHDRIFCCRSGSAADTQAIADAVTYQLGFHSIELDGPPLVHTAANLFKEMCY 121
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
R ++ G D + G VY G ++ G+ +S I +D+
Sbjct 122 RYRE-DLMAGIIVAGWDKRKGGQVYTVPMGGMLVHQQFSIGGSGSSYIYGFVDSTY---- 176
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
RP MTKEE ++F LA A ERD +G
Sbjct 177 ----RPGMTKEECLKFTANALALAMERDGSSG 204
> tgo:TGME49_114090 proteasome subunit beta type 3, putative (EC:3.4.25.1);
K02735 20S proteasome subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 34/142 (23%), Positives = 70/142 (49%), Gaps = 13/142 (9%)
Query 15 IASSGMQADINALHKWLKVRISLY--RHQHREDPSIEGLAQLLSCVLYSRRFFPYYTFNV 72
+ +G+ D+ +HK L R +LY R + E P E ++ ++S +LY RRF PY+ V
Sbjct 50 VGLAGLATDVQTVHKELVFRSNLYQLREEVTEMPP-EIMSNVVSSMLYGRRFAPYFVSPV 108
Query 73 LFGIDNQG-KGAVYGYDAVG-SFERSNYNSAGTANSLITSLLDNQIARNNQLDERPKMTK 130
+ G+ + + + +D +G + ++ GT+ + + ++ +P M +
Sbjct 109 VAGLHPETHQPFLSAFDYIGAACYAKDFVCNGTSAEQLVGVCESLW--------KPDMNE 160
Query 131 EEIIQFARGVLASAGERDIFTG 152
+E+++ L +A +RD G
Sbjct 161 DELMETLSQSLLAAVDRDCVAG 182
> dre:30666 psmb8, lmp7, macropain, prosome; proteasome (prosome,
macropain) subunit, beta type, 8 (EC:3.4.25.1); K02740 20S
proteasome subunit beta 8 [EC:3.4.25.1]
Length=271
Score = 45.8 bits (107), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 38/149 (25%), Positives = 64/149 (42%), Gaps = 9/149 (6%)
Query 4 SKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRR 63
+K+ ++ P SG AD + L LY+ ++++ S+ ++LLS ++ R
Sbjct 99 NKVIEINPYLLGTMSGSAADCQYWERLLAKECRLYKLRNKQRISVSAASKLLSNMMLGYR 158
Query 64 FFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLD 123
+++ G D QG G Y D G+ S G NS ++D+
Sbjct 159 GMGLSMGSMICGWDKQGPGLYY-VDDNGTRLSGRMFSTGCGNSYAYGVVDSGY------- 210
Query 124 ERPKMTKEEIIQFARGVLASAGERDIFTG 152
R MT EE + R +A A RD ++G
Sbjct 211 -REDMTVEEAYELGRRGIAHATHRDAYSG 238
> hsa:5696 PSMB8, D6S216, D6S216E, LMP7, MGC1491, PSMB5i, RING10;
proteasome (prosome, macropain) subunit, beta type, 8 (large
multifunctional peptidase 7) (EC:3.4.25.1); K02740 20S
proteasome subunit beta 8 [EC:3.4.25.1]
Length=272
Score = 45.4 bits (106), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 63/148 (42%), Gaps = 9/148 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ ++ P SG AD + L LY ++ E S+ ++LLS ++ R
Sbjct 101 KVIEINPYLLGTMSGCAADCQYWERLLAKECRLYYLRNGERISVSAASKLLSNMMCQYRG 160
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLDE 124
+++ G D +G G Y D G+ N S G+ N+ ++D+
Sbjct 161 MGLSMGSMICGWDKKGPGLYY-VDEHGTRLSGNMFSTGSGNTYAYGVMDSGY-------- 211
Query 125 RPKMTKEEIIQFARGVLASAGERDIFTG 152
RP ++ EE R +A A RD ++G
Sbjct 212 RPNLSPEEAYDLGRRAIAYATHRDSYSG 239
> mmu:19175 Psmb6, Lmp19, Mpnd; proteasome (prosome, macropain)
subunit, beta type 6 (EC:3.4.25.1); K02738 20S proteasome
subunit beta 1 [EC:3.4.25.1]
Length=238
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 41/152 (26%), Positives = 61/152 (40%), Gaps = 9/152 (5%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R+ KLT + SG AD A+ + ++ + + E P + A L + Y
Sbjct 62 RVTDKLTPIHDHIFCCRSGSAADTQAVADAVTYQLGFHSIELNEPPLVHTAASLFKEMCY 121
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
R ++ G D Q G VY G R ++ G+ +S I +D
Sbjct 122 RYRE-DLMAGIIIAGWDPQEGGQVYSVPMGGMMVRQSFAIGGSGSSYIYGYVDATY---- 176
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
R MTK+E +QF LA A ERD +G
Sbjct 177 ----REGMTKDECLQFTANALALAMERDGSSG 204
> hsa:5691 PSMB3, HC10-II, MGC4147; proteasome (prosome, macropain)
subunit, beta type, 3 (EC:3.4.25.1); K02735 20S proteasome
subunit beta 3 [EC:3.4.25.1]
Length=205
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/150 (22%), Positives = 67/150 (44%), Gaps = 10/150 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ + R I +G+ D+ + + LK R++LY + L +++ +LY +RF
Sbjct 41 KIFPMGDRLYIGLAGLATDVQTVAQRLKFRLNLYELKEGRQIKPYTLMSMVANLLYEKRF 100
Query 65 FPYYTFNVLFGIDNQG-KGAVYGYDAVGS-FERSNYNSAGTANSLITSLLDNQIARNNQL 122
PYYT V+ G+D + K + D +G ++ +GT + + ++
Sbjct 101 GPYYTEPVIAGLDPKTFKPFICSLDLIGCPMVTDDFVVSGTCAEQMYGMCESL------- 153
Query 123 DERPKMTKEEIIQFARGVLASAGERDIFTG 152
P M + + + + +A +RD +G
Sbjct 154 -WEPNMDPDHLFETISQAMLNAVDRDAVSG 182
> xla:734745 hypothetical protein MGC130855; K02738 20S proteasome
subunit beta 1 [EC:3.4.25.1]
Length=247
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/152 (25%), Positives = 62/152 (40%), Gaps = 9/152 (5%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R+ KLT + R SG AD A+ + ++ + + P + A L + Y
Sbjct 62 RVTDKLTPVHDRIFCCRSGSAADTQAIADAVTYQLGFHSIELEGPPLVHTAASLFKEMCY 121
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
R ++ G D + G VY G ++ G+ +S I +D+
Sbjct 122 RYRE-DLMAGIIVAGWDKRRGGQVYTVPMGGMLVHQQFSIGGSGSSYIYGFVDSTY---- 176
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
RP M+KEE ++F LA A ERD +G
Sbjct 177 ----RPGMSKEECLKFTANALALAMERDGSSG 204
> dre:30388 psmb6, y, zgc:109823; proteasome (prosome, macropain)
subunit, beta type, 6 (EC:3.4.25.1); K02738 20S proteasome
subunit beta 1 [EC:3.4.25.1]
Length=223
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 39/152 (25%), Positives = 63/152 (41%), Gaps = 9/152 (5%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R+ KLT + R SG AD A+ + ++ + + E P ++ A L + Y
Sbjct 52 RVTDKLTPIHDRIFCCRSGSAADTQAIADAVTYQLGFHSIELDEAPLVQTAASLFRDMCY 111
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
R ++ G D + G VY G R + G+ +S I +D+
Sbjct 112 RYRE-ELMAGIIVAGWDRRRGGQVYTVPVGGMLTRQPVSVGGSGSSYIYGYVDS------ 164
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
+ R M+KEE ++F G L ERD +G
Sbjct 165 --NYRSGMSKEECLKFTAGALTLPMERDGSSG 194
> ath:AT4G31300 PBA1; PBA1; endopeptidase/ peptidase/ threonine-type
endopeptidase; K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=234
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/154 (26%), Positives = 68/154 (44%), Gaps = 12/154 (7%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRH--QHREDPSIEGLAQLLSCV 58
R K+TQLT + SG AD + + VR L++H QH + +++ A L+ +
Sbjct 41 RASDKITQLTDNVYVCRSGSAADSQVVSDY--VRYFLHQHTIQHGQPATVKVSANLIRML 98
Query 59 LYSRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIAR 118
Y+ + T ++ G D G +YG G+ + G+ +S + D Q +
Sbjct 99 AYNNKQNMLQTGLIVGGWDKYEGGKIYGIPLGGTVVEQPFAIGGSGSSYLYGFFD-QAWK 157
Query 119 NNQLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
+N MTKEE Q ++ A RD +G
Sbjct 158 DN-------MTKEEAEQLVVKAVSLAIARDGASG 184
> cpv:cgd2_860 Pre3p/proteasome regulatory subunit beta type 6,
NTN hydrolase fold ; K02738 20S proteasome subunit beta 1
[EC:3.4.25.1]
Length=248
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 34/152 (22%), Positives = 69/152 (45%), Gaps = 9/152 (5%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R K+TQ+T + + SG AD + ++++ + + + ED ++ +A + + Y
Sbjct 80 RAARKITQITDKVFMCRSGSAADTQIISRYVRRIVQDHELETGEDTKVKSVASVARLISY 139
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
+ ++ G+D G+ V+ G+ +Y +G+ + I S+LD++
Sbjct 140 QNKEHLLADM-IIAGMDPNGEFKVFRIPLGGTLIEGSYAISGSGSGYIYSMLDSKYHSEM 198
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
LDE FAR +++ A RD +G
Sbjct 199 DLDECK--------SFARDLVSHAMFRDSSSG 222
> cel:K08D12.1 pbs-1; Proteasome Beta Subunit family member (pbs-1);
K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=239
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 34/134 (25%), Positives = 59/134 (44%), Gaps = 3/134 (2%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R +K+T +T + SG AD A+ K I +Y + +I +Q+ LY
Sbjct 52 RATNKITPITDNMVVCRSGSAADTQAIADIAKYHIDVYTMTENKPVTIYRSSQIFRQFLY 111
Query 61 SRRFFPYYTFNVLF-GIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARN 119
+ R + +VL G D + G VY G R ++G+ ++ + LD+Q N
Sbjct 112 NYR--EQLSASVLVAGWDEELGGQVYAIPIGGFVSRQRSTASGSGSTFVQGFLDSQWRPN 169
Query 120 NQLDERPKMTKEEI 133
L+E + K+ +
Sbjct 170 LTLEECKAIVKQAV 183
> tgo:TGME49_106930 proteasome subunit beta type 7, putative (EC:3.4.25.1);
K02739 20S proteasome subunit beta 2 [EC:3.4.25.1]
Length=368
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 36/150 (24%), Positives = 65/150 (43%), Gaps = 10/150 (6%)
Query 4 SKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRR 63
SKL ++ A +G AD++ + WL V++ L+R P + +L S+
Sbjct 166 SKLHRIADNMYAAGAGTSADLDHMCDWLAVQVELHRLNTNAKPRVSMAVSVL-----SQE 220
Query 64 FFPYYTFN----VLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARN 119
F Y + VL G+D +G +Y GS + SN+ + G+ + ++L+
Sbjct 221 LFKYQGYKGCAVVLGGVDFKGP-QIYKIHPHGSTDCSNFAAMGSGSLNAMAVLEAGYKDG 279
Query 120 NQLDERPKMTKEEIIQFARGVLASAGERDI 149
L+E + ++ I L S G D+
Sbjct 280 MTLEEGKNLVRDAIKAGVLNDLGSGGNIDL 309
> dre:64279 psmb11, MGC92422, macropain, prosome, zgc:92422; proteasome
(prosome, macropain) subunit, beta type, 11 (EC:3.4.25.1);
K02738 20S proteasome subunit beta 1 [EC:3.4.25.1]
Length=217
Score = 42.0 bits (97), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 61/148 (41%), Gaps = 9/148 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
KL Q+ R +G AD A+ K K ++S + Q P ++ A ++ + YS +
Sbjct 50 KLIQVHDRIFCCIAGSLADAQAVTKMAKFQLSFHSIQMESPPLVKAAASIMRELCYSNKE 109
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLDE 124
F + G D + +Y G + G+ ++ I +D +
Sbjct 110 ELRAGF-ITAGWDRKKGPQIYVVSLGGMLLSQPFTIGGSGSTYIYGYVDAKF-------- 160
Query 125 RPKMTKEEIIQFARGVLASAGERDIFTG 152
+P MT EE QF+ LA A RD +G
Sbjct 161 KPDMTLEEATQFSTNALALAMGRDNVSG 188
> ath:AT1G13060 PBE1; PBE1; endopeptidase/ peptidase/ threonine-type
endopeptidase; K02737 20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=274
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/129 (22%), Positives = 58/129 (44%), Gaps = 1/129 (0%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ ++ P +G AD H+ L ++ L+ ++ S+ G ++LL+ +LYS R
Sbjct 90 KIIEINPYMLGTMAGGAADCQFWHRNLGIKCRLHELANKRRISVSGASKLLANMLYSYRG 149
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLDE 124
++ G D G G Y D G + + S G+ + +LD+ + ++E
Sbjct 150 MGLSVGTMIAGWDETGPGLYY-VDNEGGRLKGDRFSVGSGSPYAYGVLDSGYKYDMSVEE 208
Query 125 RPKMTKEEI 133
++ + I
Sbjct 209 ASELARRSI 217
> ath:AT3G26340 20S proteasome beta subunit E, putative; K02737
20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=273
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/129 (22%), Positives = 58/129 (44%), Gaps = 1/129 (0%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ ++ P +G AD H+ L ++ L+ ++ S+ G ++LL+ +LYS R
Sbjct 90 KIIEINPYMLGTMAGGAADCQFWHRNLGIKCRLHELANKRRISVSGASKLLANMLYSYRG 149
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLDE 124
++ G D G G Y D G + + S G+ + +LD+ + ++E
Sbjct 150 MGLSVGTMIAGWDETGPGLYY-VDNEGGRLKGDRFSVGSGSPYAYGVLDSGYKFDMSVEE 208
Query 125 RPKMTKEEI 133
++ + I
Sbjct 209 ASELARRSI 217
> xla:378667 psmb9, LMP2, psmb9-A; proteasome (prosome, macropain)
subunit, beta type, 9 (large multifunctional peptidase
2); K02741 20S proteasome subunit beta 9 [EC:3.4.25.1]
Length=215
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 37/152 (24%), Positives = 65/152 (42%), Gaps = 10/152 (6%)
Query 1 RMHSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLY 60
R+ +KL + R A SG AD A+ + ++ + P + A L+ + Y
Sbjct 45 RVFNKLAPVHQRIYCALSGSAADAQAVADMAHYHMEVHSIEMEAPPLVLAAANLIKGISY 104
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
+ ++ G D + G VYG G R + G+ ++ I +D++
Sbjct 105 KYKE-EIMAHLIVAGWDRKHGGQVYGTLG-GMIIRQPFTIGGSGSTYIYGFVDSKF---- 158
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIFTG 152
+P M++EE +FA L+ A ERD +G
Sbjct 159 ----KPGMSREECERFAVQALSLAMERDGSSG 186
> mmu:100505005 proteasome subunit beta type-3-like
Length=205
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 31/149 (20%), Positives = 64/149 (42%), Gaps = 8/149 (5%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ + R + +G+ D+ + + LK ++LY + L +++ +LY + F
Sbjct 41 KIFPMGDRLYVGLAGLAIDVQTVAQRLKFLLNLYELKEGWQIKPYTLMSMVANLLYEKWF 100
Query 65 FPYYTFNVLFGIDNQG-KGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLD 123
PYYT V+ G+D + K + D +G T + +++ + R +
Sbjct 101 GPYYTEPVIAGLDPKTFKPFICSLDLIGC-------PMVTDDFVVSGTCSEPMYRMCESL 153
Query 124 ERPKMTKEEIIQFARGVLASAGERDIFTG 152
P M E + + + +A +RD +G
Sbjct 154 WEPNMDPEHLFETISQAMLNAVDRDAVSG 182
> mmu:73902 Psmb11, 5830406J20Rik, beta5t; proteasome (prosome,
macropain) subunit, beta type, 11 (EC:3.4.25.1); K11598 20S
proteasome subunit beta 11 [EC:3.4.25.1]
Length=302
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 36/136 (26%), Positives = 60/136 (44%), Gaps = 9/136 (6%)
Query 17 SSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRFFPYYTFNVLFGI 76
+SG AD ++ L+ + L + + PS+ G A+LL+ ++ R L G
Sbjct 94 TSGTSADCATWYRVLRRELRLRELREGQLPSVAGTAKLLAAMMSCYRGLDLCVATALCGW 153
Query 77 DNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLDERPKMTKEEIIQF 136
D+ G Y Y + G+ + + S G+ + +LD R D MT +E
Sbjct 154 DHSGPALFYVY-SDGTCLQGDIFSVGSGSPYAYGVLD----RGYHYD----MTIQEAYTL 204
Query 137 ARGVLASAGERDIFTG 152
AR +A A RD ++G
Sbjct 205 ARCAVAHATHRDAYSG 220
> mmu:16913 Psmb8, Lmp-7, Lmp7; proteasome (prosome, macropain)
subunit, beta type 8 (large multifunctional peptidase 7) (EC:3.4.25.1);
K02740 20S proteasome subunit beta 8 [EC:3.4.25.1]
Length=276
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 35/148 (23%), Positives = 61/148 (41%), Gaps = 9/148 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ ++ P SG AD + L LY ++ E S+ ++LLS ++ R
Sbjct 105 KVIEINPYLLGTMSGCAADCQYWERLLAKECRLYYLRNGERISVSAASKLLSNMMLQYRG 164
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLDE 124
+++ G D +G G Y D G+ S G+ N+ ++D+
Sbjct 165 MGLSMGSMICGWDKKGPGLYY-VDDNGTRLSGQMFSTGSGNTYAYGVMDSGY-------- 215
Query 125 RPKMTKEEIIQFARGVLASAGERDIFTG 152
R ++ EE R +A A RD ++G
Sbjct 216 RQDLSPEEAYDLGRRAIAYATHRDNYSG 243
> pfa:PF10_0111 20S proteasome beta subunit, putative; K02737
20S proteasome subunit beta 5 [EC:3.4.25.1]
Length=271
Score = 37.7 bits (86), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 31/129 (24%), Positives = 56/129 (43%), Gaps = 1/129 (0%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRF 64
K+ ++ +G AD K+L I +Y ++ E S+ + +LS +LY +
Sbjct 93 KIIEINKNILGTMAGGAADCLYWEKYLGKIIKIYELRNNEKISVRAASTILSNILYQYKG 152
Query 65 FPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLDE 124
+ +L G D+ G Y D G N S G+ ++ S+LD+ N LD+
Sbjct 153 YGLCCGIILSGYDHTGFNMFY-VDDSGKKVEGNLFSCGSGSTYAYSILDSAYDYNLNLDQ 211
Query 125 RPKMTKEEI 133
++ + I
Sbjct 212 AVELARNAI 220
> ath:AT2G05840 PAA2; PAA2 (20S PROTEASOME SUBUNIT PAA2); endopeptidase/
peptidase/ threonine-type endopeptidase; K02730 20S
proteasome subunit alpha 1 [EC:3.4.25.1]
Length=220
Score = 37.4 bits (85), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 34/149 (22%), Positives = 64/149 (42%), Gaps = 10/149 (6%)
Query 4 SKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLS--CVLYS 61
S L +T + ++GM AD +L + + + +R Q+ + + LA+ ++ +Y+
Sbjct 67 SHLFPVTKYLGLLATGMTADSRSLVQQARNEAAEFRFQYGYEMPADILAKWIADKSQVYT 126
Query 62 RRFF--PYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARN 119
+ + P ++ GID + +Y D G F SAG + L+ ++ N
Sbjct 127 QHAYMRPLGVVAMVLGIDEERGPLLYKCDPAGHFYGHKATSAGMKEQEAVNFLEKKMKEN 186
Query 120 NQLDERPKMTKEEIIQFARGVLASAGERD 148
P T +E +Q A L S + D
Sbjct 187 ------PAFTYDETVQTAISALQSVLQED 209
> hsa:122706 PSMB11, BETA5T, FLJ16369; proteasome (prosome, macropain)
subunit, beta type, 11 (EC:3.4.25.1); K11598 20S proteasome
subunit beta 11 [EC:3.4.25.1]
Length=300
Score = 37.0 bits (84), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 59/136 (43%), Gaps = 9/136 (6%)
Query 17 SSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVLYSRRFFPYYTFNVLFGI 76
+SG AD ++ L+ + L + + PS+ A+LLS ++ R L G
Sbjct 94 TSGTSADCATWYRVLQRELRLRELREGQLPSVASAAKLLSAMMSQYRGLDLCVATALCGW 153
Query 77 DNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQLDERPKMTKEEIIQF 136
D G Y Y + G+ + + S G+ + +LD R + D M+ +E
Sbjct 154 DRSGPELFYVY-SDGTRLQGDIFSVGSGSPYAYGVLD----RGYRYD----MSTQEAYAL 204
Query 137 ARGVLASAGERDIFTG 152
AR +A A RD ++G
Sbjct 205 ARCAVAHATHRDAYSG 220
> dre:436882 psmb2, zgc:92282; proteasome (prosome, macropain)
subunit, beta type, 2 (EC:3.4.25.1); K02734 20S proteasome
subunit beta 4 [EC:3.4.25.1]
Length=199
Score = 35.8 bits (81), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 31/150 (20%), Positives = 61/150 (40%), Gaps = 10/150 (6%)
Query 3 HSKLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVL--Y 60
+ K+ +L+ + + G D ++++ + LY+ ++ + S A L Y
Sbjct 32 YDKMFKLSEKILLLCVGEAGDTVQFAEYIQKNVQLYKMRNGYELSPAAAANFTRKNLADY 91
Query 61 SRRFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNN 120
R PY+ +L G D +Y D + + ++ + + G L S+LD
Sbjct 92 LRSRTPYHVNLLLAGYDETDGPGLYYMDYLSALAKAPFAAHGYGAFLTLSILDRYY---- 147
Query 121 QLDERPKMTKEEIIQFARGVLASAGERDIF 150
RP +T+EE + + L +R I
Sbjct 148 ----RPDLTREEAVDLLKKCLEELNKRFIL 173
> xla:443966 psmb2, MGC130720, MGC80364; proteasome (prosome,
macropain) subunit, beta type, 2; K02734 20S proteasome subunit
beta 4 [EC:3.4.25.1]
Length=199
Score = 35.8 bits (81), Expect = 0.057, Method: Compositional matrix adjust.
Identities = 29/149 (19%), Positives = 64/149 (42%), Gaps = 10/149 (6%)
Query 5 KLTQLTPRCSIASSGMQADINALHKWLKVRISLYRHQHREDPSIEGLAQLLSCVL--YSR 62
K+ +++ + + G D ++++ + LY+ ++ + S A L Y R
Sbjct 34 KMFKMSEKILLLCVGEAGDTVQFAEYIQKNVQLYKMRNGYELSPTAAANFTRRNLADYLR 93
Query 63 RFFPYYTFNVLFGIDNQGKGAVYGYDAVGSFERSNYNSAGTANSLITSLLDNQIARNNQL 122
PY+ +L G D ++Y D + + ++ + + G L S+LD
Sbjct 94 SRTPYHVNLLLAGYDEHEGPSLYYMDYLAALAKTRFAAHGYGAYLTLSILDRYY------ 147
Query 123 DERPKMTKEEIIQFARGVLASAGERDIFT 151
+P +T+EE ++ + +A +R I +
Sbjct 148 --KPDLTREEAVELLKKCIAELQKRFILS 174
Lambda K H
0.320 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40