bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2910_orf1
Length=176
Score E
Sequences producing significant alignments: (Bits) Value
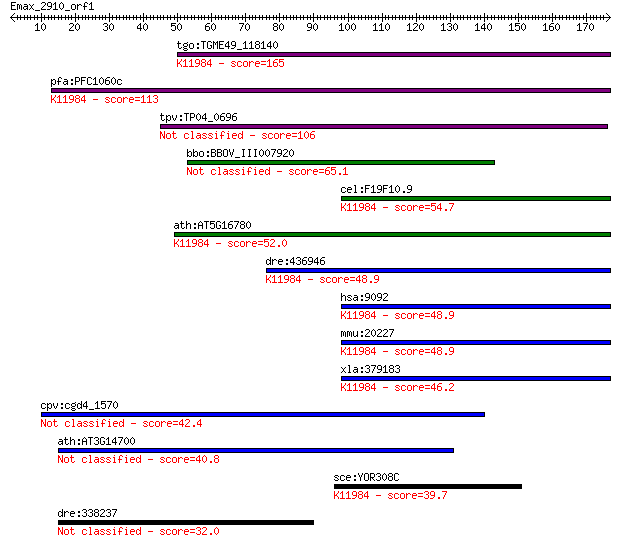
tgo:TGME49_118140 hypothetical protein ; K11984 U4/U6.U5 tri-s... 165 7e-41
pfa:PFC1060c conserved Plasmodium protein, unknown function; K... 113 4e-25
tpv:TP04_0696 hypothetical protein 106 3e-23
bbo:BBOV_III007920 17.m07694; hypothetical protein 65.1 1e-10
cel:F19F10.9 hypothetical protein; K11984 U4/U6.U5 tri-snRNP-a... 54.7 2e-07
ath:AT5G16780 DOT2; DOT2 (DEFECTIVELY ORGANIZED TRIBUTARIES 2)... 52.0 1e-06
dre:436946 sart1, zgc:91927; squamous cell carcinoma antigen r... 48.9 8e-06
hsa:9092 SART1, Ara1, HOMS1, MGC2038, SART1259, SNRNP110, Snu6... 48.9 9e-06
mmu:20227 Sart1, U5-110K; squamous cell carcinoma antigen reco... 48.9 9e-06
xla:379183 sart1, MGC132129, MGC53679; squamous cell carcinoma... 46.2 6e-05
cpv:cgd4_1570 hypothetical protein 42.4 8e-04
ath:AT3G14700 hypothetical protein 40.8 0.002
sce:YOR308C SNU66; Component of the U4/U6.U5 snRNP complex inv... 39.7 0.005
dre:338237 id:ibd1338; si:ch211-266d19.3 32.0 1.1
> tgo:TGME49_118140 hypothetical protein ; K11984 U4/U6.U5 tri-snRNP-associated
protein 1
Length=861
Score = 165 bits (418), Expect = 7e-41, Method: Compositional matrix adjust.
Identities = 75/127 (59%), Positives = 101/127 (79%), Gaps = 0/127 (0%)
Query 50 FMAEQPIGDSVSGALQYLQSKDFFSLDKIRRRRGHHLEQPLHNADNDKEVNIDYRDAFGN 109
FM E P+G ++ AL+YLQSK+ +SLDK+R+RR E PLH +K++ ID+RD +GN
Sbjct 729 FMNENPMGHGLAEALKYLQSKNHYSLDKMRQRRHRPDELPLHKPLGEKDIKIDHRDQYGN 788
Query 110 VMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIMDCLPTLKALKKVQEGES 169
VMT KDAFR ISW FHGK+PSLRKQEKK+KK+++ER+L +N M+ LPTL AL+++QE E
Sbjct 789 VMTPKDAFREISWRFHGKYPSLRKQEKKMKKMDIERKLLQNPMEALPTLSALQRLQEKEK 848
Query 170 SAHLVLT 176
++HLVLT
Sbjct 849 ASHLVLT 855
> pfa:PFC1060c conserved Plasmodium protein, unknown function;
K11984 U4/U6.U5 tri-snRNP-associated protein 1
Length=693
Score = 113 bits (282), Expect = 4e-25, Method: Compositional matrix adjust.
Identities = 66/169 (39%), Positives = 104/169 (61%), Gaps = 8/169 (4%)
Query 13 SSSSKKQREETEEGEVVENVPKEDEEEEEEEGDSE----PEFMAEQPIGDSVSGALQYLQ 68
+SS+K EE ++++N E+EE+ ++ SE E E + + + GAL+YL+
Sbjct 525 NSSNKNILEENINEDILKNTFLENEEDHNDDNSSELHGVSEIFNEVKLDEGLFGALEYLK 584
Query 69 SKDFFSL-DKIRRRRGHHLEQPLHNADNDKEVNIDYRDAFGNVMTAKDAFRRISWHFHGK 127
+K ++ DKI R + +PLH + + ++ +DY++ FG VMT K++FR ISW FHGK
Sbjct 585 TKGELNMEDKIYRNPEN---KPLHMSTDKDDIKLDYKNEFGKVMTPKESFRYISWIFHGK 641
Query 128 FPSLRKQEKKIKKLELERRLQENIMDCLPTLKALKKVQEGESSAHLVLT 176
K EKKIK+LE+ERR +EN +D LPTL LKK Q+ + ++ L+
Sbjct 642 KQGKNKLEKKIKRLEIERRYKENPIDSLPTLNVLKKYQQTQKKSYFTLS 690
> tpv:TP04_0696 hypothetical protein
Length=554
Score = 106 bits (265), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 54/131 (41%), Positives = 81/131 (61%), Gaps = 14/131 (10%)
Query 45 DSEPEFMAEQPIGDSVSGALQYLQSKDFFSLDKIRRRRGHHLEQPLHNADNDKEVNIDYR 104
D +P ++EQP+GD ++ AL Y+ ++RG ++++ KEV ++Y
Sbjct 431 DDDPNTLSEQPLGDGIAAALSYI------------KQRGDYIDEKAETRS--KEVQLNYL 476
Query 105 DAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIMDCLPTLKALKKV 164
D +GN MT K+AF++ISW FHGK PS +KQEK +K+ELER L N + LPT+KAL
Sbjct 477 DEYGNEMTPKEAFKKISWIFHGKRPSKKKQEKMRRKIELERALNSNPVGGLPTMKALYSH 536
Query 165 QEGESSAHLVL 175
QE E + ++ L
Sbjct 537 QEKEQTPYITL 547
> bbo:BBOV_III007920 17.m07694; hypothetical protein
Length=528
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 53/90 (58%), Gaps = 12/90 (13%)
Query 53 EQPIGDSVSGALQYLQSKDFFSLDKIRRRRGHHLEQPLHNADNDKEVNIDYRDAFGNVMT 112
++P+ ++GAL YL+ K D I +++ L ND + + Y D +G MT
Sbjct 439 DEPMTTGIAGALAYLKDKG----DIIEKKKD------LEGVGND--ITLQYFDEYGRKMT 486
Query 113 AKDAFRRISWHFHGKFPSLRKQEKKIKKLE 142
K+AFR++SW FHGK P L K+E+ IK++E
Sbjct 487 PKEAFRQLSWKFHGKGPGLNKRERIIKRIE 516
> cel:F19F10.9 hypothetical protein; K11984 U4/U6.U5 tri-snRNP-associated
protein 1
Length=829
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/81 (41%), Positives = 50/81 (61%), Gaps = 2/81 (2%)
Query 98 EVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKI-KKLELERRLQENIMDC-L 155
+VNI Y D G M AKDA+R +S+ FHG+ P ++ EK+ +K + ER L+ N D L
Sbjct 738 DVNISYVDRKGREMDAKDAYRELSYKFHGRNPGKKQLEKRANRKDKEERMLKTNSYDTPL 797
Query 156 PTLKALKKVQEGESSAHLVLT 176
TL +K Q+ S+ +LVL+
Sbjct 798 GTLDKQRKKQKQLSTPYLVLS 818
> ath:AT5G16780 DOT2; DOT2 (DEFECTIVELY ORGANIZED TRIBUTARIES
2); K11984 U4/U6.U5 tri-snRNP-associated protein 1
Length=820
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 39/146 (26%), Positives = 74/146 (50%), Gaps = 18/146 (12%)
Query 49 EFMAEQPIGDSVSGALQYLQSKDFF---------SLDKIRRR-------RGHHLEQPLHN 92
E + E +G +SGAL+ L+ + ++DK + + G + +
Sbjct 616 ENIHEVAVGKGLSGALKLLKDRGTLKEKVEWGGRNMDKKKSKLVGIVDDDGGKESKDKES 675
Query 93 ADNDKEVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIM 152
D K++ I+ D FG +T K+AFR +S FHGK P K+EK++K+ + E +L++
Sbjct 676 KDRFKDIRIERTDEFGRTLTPKEAFRLLSHKFHGKGPGKMKEEKRMKQYQEELKLKQMKN 735
Query 153 DCLP--TLKALKKVQEGESSAHLVLT 176
P +++ +++ Q + +LVL+
Sbjct 736 SDTPSQSVQRMREAQAQLKTPYLVLS 761
> dre:436946 sart1, zgc:91927; squamous cell carcinoma antigen
recognised by T cells; K11984 U4/U6.U5 tri-snRNP-associated
protein 1
Length=777
Score = 48.9 bits (115), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 36/105 (34%), Positives = 57/105 (54%), Gaps = 4/105 (3%)
Query 76 DKIRRRRGHH-LEQPLHNADNDK-EVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRK 133
DK RR + Q DN K +V I+Y D G + K+AFR++S FHGK K
Sbjct 660 DKYSRREEYRGFTQDFKEKDNYKPDVKIEYVDESGRKLCPKEAFRQLSHRFHGKGSGKMK 719
Query 134 QEKKIKKLELERRLQENIMDCLP--TLKALKKVQEGESSAHLVLT 176
E+++KKLE E L++ P T+ L++ Q+ + + ++VL+
Sbjct 720 TERRMKKLEEEALLKKMSSSDTPLGTVALLQEKQKSQKTPYIVLS 764
> hsa:9092 SART1, Ara1, HOMS1, MGC2038, SART1259, SNRNP110, Snu66;
squamous cell carcinoma antigen recognized by T cells;
K11984 U4/U6.U5 tri-snRNP-associated protein 1
Length=800
Score = 48.9 bits (115), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 49/81 (60%), Gaps = 2/81 (2%)
Query 98 EVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIMDCLP- 156
+V I+Y D G +T K+AFR++S FHGK K E+++KKL+ E L++ P
Sbjct 707 DVKIEYVDETGRKLTPKEAFRQLSHRFHGKGSGKMKTERRMKKLDEEALLKKMSSSDTPL 766
Query 157 -TLKALKKVQEGESSAHLVLT 176
T+ L++ Q+ + + ++VL+
Sbjct 767 GTVALLQEKQKAQKTPYIVLS 787
> mmu:20227 Sart1, U5-110K; squamous cell carcinoma antigen recognized
by T-cells 1; K11984 U4/U6.U5 tri-snRNP-associated
protein 1
Length=806
Score = 48.9 bits (115), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 28/81 (34%), Positives = 49/81 (60%), Gaps = 2/81 (2%)
Query 98 EVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIMDCLP- 156
+V I+Y D G +T K+AFR++S FHGK K E+++KKL+ E L++ P
Sbjct 713 DVKIEYVDETGRKLTPKEAFRQLSHRFHGKGSGKMKTERRMKKLDEEALLKKMSSSDTPL 772
Query 157 -TLKALKKVQEGESSAHLVLT 176
T+ L++ Q+ + + ++VL+
Sbjct 773 GTVALLQEKQKAQKTPYIVLS 793
> xla:379183 sart1, MGC132129, MGC53679; squamous cell carcinoma
antigen recognized by T cells; K11984 U4/U6.U5 tri-snRNP-associated
protein 1
Length=765
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 48/81 (59%), Gaps = 2/81 (2%)
Query 98 EVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQENIMDCLP- 156
+V I+Y D G + K+AFR++S FHGK K E+++KKL+ E L++ P
Sbjct 672 DVKIEYVDETGRKLCPKEAFRQLSHRFHGKGSGKMKTERRMKKLDEEALLKKMSSSDTPL 731
Query 157 -TLKALKKVQEGESSAHLVLT 176
T+ L++ Q+ + + ++VL+
Sbjct 732 GTVALLQEKQKAQKTPYIVLS 752
> cpv:cgd4_1570 hypothetical protein
Length=407
Score = 42.4 bits (98), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 32/141 (22%), Positives = 69/141 (48%), Gaps = 20/141 (14%)
Query 10 SSSSSSSKKQREETEEGEVVENVPKEDEEEEEEEGDSEPEFMAEQPIGDSVSGALQYLQS 69
SS+ ++ +K + TEE V +E+ + + + E+P+ +S L+ L+
Sbjct 269 SSNDNNRQKTSKNTEETRV---------SDEKTQNNLNNNILYEEPLDFGISSTLELLKK 319
Query 70 KDFFSL-----------DKIRRRRGHHLEQPLHNADNDKEVNIDYRDAFGNVMTAKDAFR 118
+ S ++ ++ ++ N+++D +V+I + D GN++ K+AF+
Sbjct 320 RGNISSSNKKDPITSNNNEFGQKNENYSTDSALNSESDFQVSILHTDDNGNILNPKEAFK 379
Query 119 RISWHFHGKFPSLRKQEKKIK 139
R+ W FHG+ + K EK ++
Sbjct 380 RLCWKFHGQKVNKNKIEKMLR 400
> ath:AT3G14700 hypothetical protein
Length=204
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 57/122 (46%), Gaps = 17/122 (13%)
Query 15 SSKKQREETEEGEVVENVPKEDEEEEEEEGDSEPEFMAEQPIGDSVSGALQYLQSKDFFS 74
SS+++RE + E + V K + GD M E +G +SGAL L+ + F
Sbjct 52 SSERRREVCSKAEDI--VDKAIDNHSRVRGDG---IMREADVGTGLSGALNRLREQGTF- 105
Query 75 LDKIRRRRGHHLEQPLHNADND------KEVNIDYRDAFGNVMTAKDAFRRISWHFHGKF 128
+ G + +N ++D K++ I + +G +MT K+A+R + FHGK
Sbjct 106 -----KEEGKVVGVKDNNHEDDRFKDRFKDIQIQRVNKWGRIMTEKEAYRSLCHGFHGKG 160
Query 129 PS 130
P
Sbjct 161 PG 162
> sce:YOR308C SNU66; Component of the U4/U6.U5 snRNP complex involved
in pre-mRNA splicing via spliceosome; also required
for pre-5S rRNA processing and may act in concert with Rnh70p;
has homology to human SART-1; K11984 U4/U6.U5 tri-snRNP-associated
protein 1
Length=587
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 33/55 (60%), Gaps = 0/55 (0%)
Query 96 DKEVNIDYRDAFGNVMTAKDAFRRISWHFHGKFPSLRKQEKKIKKLELERRLQEN 150
D ++ + YRD GN +T K+A++++S FHG + +K+ K ++E + EN
Sbjct 524 DPDIKLVYRDEKGNRLTTKEAYKKLSQKFHGTKSNKKKRAKMKSRIEARKNTPEN 578
> dre:338237 id:ibd1338; si:ch211-266d19.3
Length=1677
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 30/80 (37%), Positives = 43/80 (53%), Gaps = 5/80 (6%)
Query 15 SSKKQREETE-EGEVVENVPKEDEEE--EEEEGDSEPEFMAEQPIGDS-VSGALQYLQSK 70
S +K + ETE G VVE+ P ED+EE +EEE +EP+ +P + + +K
Sbjct 1446 SVRKGKAETEGNGSVVESGPDEDKEERSDEEEPATEPKSAGREPGSKPDKRKKVCSICNK 1505
Query 71 DFFSL-DKIRRRRGHHLEQP 89
F+SL D R R H E+P
Sbjct 1506 RFWSLQDLTRHMRSHTGERP 1525
Lambda K H
0.308 0.127 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4600750868
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40