bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2931_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
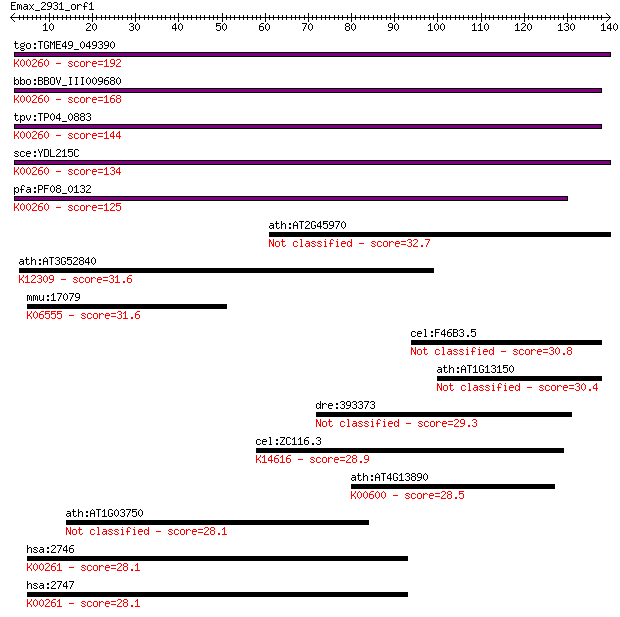
tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putati... 192 3e-49
bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/... 168 4e-42
tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2... 144 6e-35
sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydro... 134 7e-32
pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2); ... 125 5e-29
ath:AT2G45970 CYP86A8; fatty acid (omega-1)-hydroxylase/ oxyge... 32.7 0.31
ath:AT3G52840 BGAL2; BGAL2 (beta-galactosidase 2); beta-galact... 31.6 0.82
mmu:17079 Cd180, F630107B15, Ly78, RP105; CD180 antigen; K0655... 31.6 0.85
cel:F46B3.5 grd-2; GRounDhog (hedgehog-like family) family mem... 30.8 1.5
ath:AT1G13150 CYP86C4; electron carrier/ heme binding / iron i... 30.4 1.7
dre:393373 MGC63882; zgc:63882 29.3 3.6
cel:ZC116.3 hypothetical protein; K14616 cubilin 28.9 5.1
ath:AT4G13890 EDA36; EDA36 (EMBRYO SAC DEVELOPMENT ARREST 37);... 28.5 6.4
ath:AT1G03750 SWI2; SWI2 (SWITCH 2); ATP binding / DNA binding... 28.1 8.2
hsa:2746 GLUD1, GDH, GDH1, GLUD, MGC132003; glutamate dehydrog... 28.1 8.4
hsa:2747 GLUD2, GDH2, GLUDP1; glutamate dehydrogenase 2 (EC:1.... 28.1 9.4
> tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putative
(EC:1.4.1.2); K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1113
Score = 192 bits (488), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 89/138 (64%), Positives = 111/138 (80%), Gaps = 0/138 (0%)
Query 2 WWKAFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGC 61
+WKAFTTGKL GGIPHD +GMTTAS+E ++ GI K NLKEEE+TR GGPDGDLG
Sbjct 716 FWKAFTTGKLPAMGGIPHDTYGMTTASIETYIHGILEKKNLKEEEVTRQLVGGPDGDLGS 775
Query 62 NALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGFK 121
NALL++ +KT +++D SGVL+DP+GL+ EL RL + RFEG +T+AMLY+ LLSP GFK
Sbjct 776 NALLKSNTKTTSIVDGSGVLHDPEGLDINELRRLAKRRFEGLQTSAMLYDEKLLSPMGFK 835
Query 122 VAQDARDIILPDGTFVAS 139
V+QD RD++LPDGT VAS
Sbjct 836 VSQDDRDVVLPDGTAVAS 853
> bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/Valine
dehydrogenase family protein (EC:1.4.1.2); K00260 glutamate
dehydrogenase [EC:1.4.1.2]
Length=1025
Score = 168 bits (426), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 81/136 (59%), Positives = 104/136 (76%), Gaps = 0/136 (0%)
Query 2 WWKAFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGC 61
+W++FTTGK GGIPHD +GMTTAS+EA++ + N +L EEE+TR TGGPDGDLG
Sbjct 643 FWRSFTTGKEPVMGGIPHDTYGMTTASIEAYIHELLNIFHLNEEEVTRFLTGGPDGDLGS 702
Query 62 NALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGFK 121
NALL +K+KT+ VID SGVL+DP+GL+ EL RL R +G T+AM YN +LLS +GFK
Sbjct 703 NALLCSKTKTLTVIDKSGVLHDPEGLDINELQRLAANRLKGLPTSAMHYNEALLSDKGFK 762
Query 122 VAQDARDIILPDGTFV 137
V +DA D++LPDGT V
Sbjct 763 VPEDAVDMVLPDGTKV 778
> tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1178
Score = 144 bits (364), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 79/172 (45%), Positives = 101/172 (58%), Gaps = 36/172 (20%)
Query 2 WWKAFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGC 61
+W++FTTGK Q GGIPHD +GMTT S+EA+V GI NK LKEEE+TR TGGPDGDLG
Sbjct 731 YWRSFTTGKAPQLGGIPHDIYGMTTTSIEAYVTGILNKYGLKEEEVTRFLTGGPDGDLGS 790
Query 62 NALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLR---------------------- 99
NA+ + +KT+ V+D SGVL+DP GL+ EL RL LR
Sbjct 791 NAIKVSNTKTLTVLDKSGVLHDPNGLDLNELRRLAFLRDTTHLSATDNPNNVTLNHSNSS 850
Query 100 ------FEGKE--------TNAMLYNSSLLSPQGFKVAQDARDIILPDGTFV 137
EG E T +M Y+ LLS +GF V ++A +++LPDG V
Sbjct 851 LQRSSSLEGDEELLGARLKTCSMGYDKRLLSSKGFMVPEEAMNVVLPDGFVV 902
> sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydrogenase
[EC:1.4.1.2]
Length=1092
Score = 134 bits (338), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 71/140 (50%), Positives = 92/140 (65%), Gaps = 7/140 (5%)
Query 2 WWKAFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGC 61
WWK+F TGK GGIPHD +GMT+ V A+V IY LNL + + QTGGPDGDLG
Sbjct 701 WWKSFLTGKSPSLGGIPHDEYGMTSLGVRAYVNKIYETLNLTNSTVYKFQTGGPDGDLGS 760
Query 62 NALLQTKSKT--IAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQG 119
N +L + +A++D SGVL DPKGL+K+E LCRL E K + +++S LS G
Sbjct 761 NEILLSSPNECYLAILDGSGVLCDPKGLDKDE---LCRLAHERKMISD--FDTSKLSNNG 815
Query 120 FKVAQDARDIILPDGTFVAS 139
F V+ DA DI+LP+GT VA+
Sbjct 816 FFVSVDAMDIMLPNGTIVAN 835
> pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1397
Score = 125 bits (313), Expect = 5e-29, Method: Composition-based stats.
Identities = 63/132 (47%), Positives = 93/132 (70%), Gaps = 4/132 (3%)
Query 2 WWKAFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGC 61
+WK F+TGKL ++GG+PHD +GMTT +E ++ + KLN+KEE ++R GGPDGDLG
Sbjct 932 YWKTFSTGKLRKNGGVPHDMYGMTTLGIETYISKLCEKLNIKEESISRSLVGGPDGDLGS 991
Query 62 NALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAM----LYNSSLLSP 117
NA+LQ+K+K I++ID SG+LYD +GLNKEEL RL + R ++ A+ LY+ S
Sbjct 992 NAILQSKTKIISIIDGSGILYDKQGLNKEELIRLAKRRNNKDKSKAITCCTLYDEKYFSK 1051
Query 118 QGFKVAQDARDI 129
GFK++ + ++
Sbjct 1052 DGFKISIEDHNV 1063
> ath:AT2G45970 CYP86A8; fatty acid (omega-1)-hydroxylase/ oxygen
binding
Length=537
Score = 32.7 bits (73), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 40/79 (50%), Gaps = 13/79 (16%)
Query 61 CNALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGF 120
C L++T+ +A L+ + L+ EEL RL L+ ET LY S P+
Sbjct 336 CTVLVETRGDDVA-------LWTDEPLSCEELDRLVFLKAALSET-LRLYPSV---PEDS 384
Query 121 KVAQDARDIILPDGTFVAS 139
K A +D +LPDGTFV +
Sbjct 385 KRA--VKDDVLPDGTFVPA 401
> ath:AT3G52840 BGAL2; BGAL2 (beta-galactosidase 2); beta-galactosidase/
catalytic/ cation binding; K12309 beta-galactosidase
[EC:3.2.1.23]
Length=727
Score = 31.6 bits (70), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 42/97 (43%), Gaps = 7/97 (7%)
Query 3 WKAFTTGKLIQHGG-IPHDRFGMTTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGC 61
W TG + GG IP+ SV F++ + +N + GG + D
Sbjct 252 WTENWTGWFTEFGGAIPNRPVEDIAFSVARFIQNGGSFMNY------YMYYGGTNFDRTA 305
Query 62 NALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRL 98
+ T A ID G+L +PK + +ELH++ +L
Sbjct 306 GVFIATSYDYDAPIDEYGLLREPKYSHLKELHKVIKL 342
> mmu:17079 Cd180, F630107B15, Ly78, RP105; CD180 antigen; K06555
lymphocyte antigen 78
Length=661
Score = 31.6 bits (70), Expect = 0.85, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 31/48 (64%), Gaps = 6/48 (12%)
Query 5 AFTTGKLIQHGGIPHDRFGMTTASVEAF--VKGIYNKLNLKEEEMTRI 50
AFT+ K++ H + H+R +T++S+EA +KGIY LNL ++ I
Sbjct 515 AFTSLKMMNHVDLSHNR--LTSSSIEALSHLKGIY--LNLASNRISII 558
> cel:F46B3.5 grd-2; GRounDhog (hedgehog-like family) family member
(grd-2)
Length=946
Score = 30.8 bits (68), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 22/44 (50%), Gaps = 2/44 (4%)
Query 94 RLCRLRFEGKETNAMLYNSSLLSPQGFKVAQDARDIILPDGTFV 137
R+C++ EG NA+ Y SSL P+ D+ L TFV
Sbjct 610 RMCKIDHEG--YNALTYQSSLRPPRAIDFIDFPNDVTLDGPTFV 651
> ath:AT1G13150 CYP86C4; electron carrier/ heme binding / iron
ion binding / monooxygenase/ oxygen binding
Length=529
Score = 30.4 bits (67), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 23/45 (51%), Gaps = 7/45 (15%)
Query 100 FEGKETNAMLYNSSLLS-------PQGFKVAQDARDIILPDGTFV 137
F KE N M+Y + LS P ++ Q D +LPDGTFV
Sbjct 365 FTVKELNNMVYLQAALSETLRLFPPIPMEMKQAIEDDVLPDGTFV 409
> dre:393373 MGC63882; zgc:63882
Length=306
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 16/62 (25%), Positives = 31/62 (50%), Gaps = 3/62 (4%)
Query 72 IAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYN---SSLLSPQGFKVAQDARD 128
+A + + P GL KE+L +CR EG+E + + SS P+G +++ ++
Sbjct 80 LAALGHKTIKRQPTGLTKEDLADVCRREGEGEERDVDRVSGLGSSRSGPRGMPLSRQEKE 139
Query 129 II 130
+
Sbjct 140 AV 141
> cel:ZC116.3 hypothetical protein; K14616 cubilin
Length=4047
Score = 28.9 bits (63), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 18/71 (25%), Positives = 30/71 (42%), Gaps = 5/71 (7%)
Query 58 DLGCNALLQTKSKTIAVIDASGVLYDPKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSP 117
D C L+ ++ ++ D K +NKE H+ CR +G +T ++ N
Sbjct 3642 DSDCGGWLKATNEIKTLVYKGITSDDNKEMNKERSHQRCRFMIQGPKTEPVIVNF----- 3696
Query 118 QGFKVAQDARD 128
Q F + A D
Sbjct 3697 QQFNIPSKAGD 3707
> ath:AT4G13890 EDA36; EDA36 (EMBRYO SAC DEVELOPMENT ARREST 37);
catalytic/ glycine hydroxymethyltransferase/ pyridoxal phosphate
binding (EC:2.1.2.1); K00600 glycine hydroxymethyltransferase
[EC:2.1.2.1]
Length=470
Score = 28.5 bits (62), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 14/49 (28%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 80 VLYD--PKGLNKEELHRLCRLRFEGKETNAMLYNSSLLSPQGFKVAQDA 126
+L+D P GL ++ ++C L + NA+ ++S L+P G ++ A
Sbjct 346 ILWDLRPLGLTGNKVEKVCELCYITLNRNAVFGDTSFLAPGGVRIGTPA 394
> ath:AT1G03750 SWI2; SWI2 (SWITCH 2); ATP binding / DNA binding
/ helicase/ nucleic acid binding
Length=862
Score = 28.1 bits (61), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 31/71 (43%), Gaps = 17/71 (23%)
Query 14 HGGIPHDRFGM-TTASVEAFVKGIYNKLNLKEEEMTRIQTGGPDGDLGCNALLQTKSKTI 72
HGGI D G+ T AF+ +Y G DGD G + LL++ +
Sbjct 158 HGGILGDDMGLGKTIQTIAFLAAVY----------------GKDGDAGESCLLESDKGPV 201
Query 73 AVIDASGVLYD 83
+I S ++++
Sbjct 202 LIICPSSIIHN 212
> hsa:2746 GLUD1, GDH, GDH1, GLUD, MGC132003; glutamate dehydrogenase
1 (EC:1.4.1.3); K00261 glutamate dehydrogenase (NAD(P)+)
[EC:1.4.1.3]
Length=558
Score = 28.1 bits (61), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 43/97 (44%), Gaps = 10/97 (10%)
Query 5 AFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLKE---------EEMTRIQTGGP 55
A TGK I GGI H R T V ++ N+ + ++ +Q G
Sbjct 253 ACVTGKPISQGGI-HGRISATGRGVFHGIENFINEASYMSILGMTPGFGDKTFVVQGFGN 311
Query 56 DGDLGCNALLQTKSKTIAVIDASGVLYDPKGLNKEEL 92
G L + +K IAV ++ G +++P G++ +EL
Sbjct 312 VGLHSMRYLHRFGAKCIAVGESDGSIWNPDGIDPKEL 348
> hsa:2747 GLUD2, GDH2, GLUDP1; glutamate dehydrogenase 2 (EC:1.4.1.3);
K00261 glutamate dehydrogenase (NAD(P)+) [EC:1.4.1.3]
Length=558
Score = 28.1 bits (61), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 43/97 (44%), Gaps = 10/97 (10%)
Query 5 AFTTGKLIQHGGIPHDRFGMTTASVEAFVKGIYNKLNLK---------EEEMTRIQTGGP 55
A TGK I GGI H R T V ++ N+ + ++ +Q G
Sbjct 253 ACVTGKPISQGGI-HGRISATGRGVFHGIENFINEASYMSILGMTPGFRDKTFVVQGFGN 311
Query 56 DGDLGCNALLQTKSKTIAVIDASGVLYDPKGLNKEEL 92
G L + +K IAV ++ G +++P G++ +EL
Sbjct 312 VGLHSMRYLHRFGAKCIAVGESDGSIWNPDGIDPKEL 348
Lambda K H
0.318 0.136 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2487377096
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40