bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_2999_orf2
Length=67
Score E
Sequences producing significant alignments: (Bits) Value
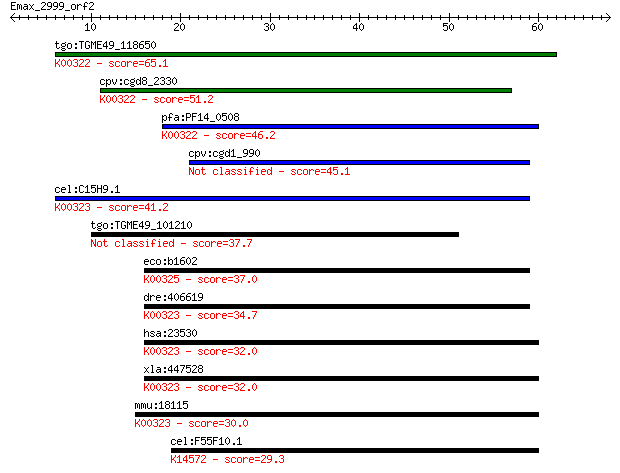
tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00... 65.1 5e-11
cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alp... 51.2 8e-07
pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative (... 46.2 2e-05
cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alph... 45.1 5e-05
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 41.2 8e-04
tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, puta... 37.7 0.010
eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydr... 37.0 0.014
dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamid... 34.7 0.067
hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide t... 32.0 0.48
xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydroge... 32.0 0.54
mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide... 30.0 1.9
cel:F55F10.1 hypothetical protein; K14572 midasin 29.3 3.5
> tgo:TGME49_118650 transhydrogenase, putative (EC:1.6.1.2); K00322
NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1013
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 32/56 (57%), Positives = 45/56 (80%), Gaps = 2/56 (3%)
Query 6 AEGVGAAAPQGAVQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAKVSA 61
AEGV +G V++ + D VA+E+L A+KVLIVPGYGMAV+RC+S++ADIA++ A
Sbjct 275 AEGVPGV--EGVVREISPDSVAEEVLLAKKVLIVPGYGMAVSRCQSDVADIAQILA 328
> cpv:cgd8_2330 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, possible signal peptide
plus 12 transmembrane regions ; K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=1143
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 25/46 (54%), Positives = 34/46 (73%), Gaps = 0/46 (0%)
Query 11 AAAPQGAVQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADI 56
A Q V +T A +VA +LL+A+KVLIVPGYGMAV+R + ++A I
Sbjct 372 ALGDQSDVNKTNAMKVARDLLSAKKVLIVPGYGMAVSRSQQDVASI 417
> pfa:PF14_0508 pyridine nucleotide transhydrogenase, putative
(EC:1.6.1.2); K00322 NAD(P) transhydrogenase [EC:1.6.1.1]
Length=1176
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 30/42 (71%), Gaps = 0/42 (0%)
Query 18 VQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAKV 59
+ TT VA+ L+ A+ ++IVPGYG A+++C+ ELA+I +
Sbjct 389 INSTTNKYVAENLINAKNIIIVPGYGTALSKCQRELAEICSI 430
> cpv:cgd1_990 pyridine nucleotide/ NAD(P) transhydrogenase alpha
plus beta subunits, duplicated gene, 12 transmembrane domain
(EC:1.6.1.2)
Length=1147
Score = 45.1 bits (105), Expect = 5e-05, Method: Composition-based stats.
Identities = 19/38 (50%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 21 TTADEVADELLAARKVLIVPGYGMAVARCRSELADIAK 58
T++ E A+ L + K+LIVPGYGMAV++C+ ++DI K
Sbjct 345 TSSKETAEILAESSKILIVPGYGMAVSKCQDIISDIIK 382
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 21/54 (38%), Positives = 33/54 (61%), Gaps = 1/54 (1%)
Query 6 AEGVG-AAAPQGAVQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAK 58
++G G A A +G ++ E AD LL AR V+I+PGYG+ A+ + +A + K
Sbjct 852 SKGTGEAKAIEGTAKEIAPVETADMLLNARSVIIIPGYGLCAAQAQYPIAQLVK 905
> tgo:TGME49_101210 NAD(P) transhydrogenase, alpha subunit, putative
(EC:1.6.1.2)
Length=1165
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 29/41 (70%), Gaps = 0/41 (0%)
Query 10 GAAAPQGAVQQTTADEVADELLAARKVLIVPGYGMAVARCR 50
A A +G + T+++VA LLA++ V+IVPGYGMAV+ +
Sbjct 268 NAQAFEGEAKIATSEQVAAYLLASQSVIIVPGYGMAVSHAQ 308
> eco:b1602 pntB, ECK1597, JW1594; pyridine nucleotide transhydrogenase,
beta subunit (EC:1.6.1.2); K00325 NAD(P) transhydrogenase
subunit beta [EC:1.6.1.2]
Length=462
Score = 37.0 bits (84), Expect = 0.014, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 29/43 (67%), Gaps = 0/43 (0%)
Query 16 GAVQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAK 58
G ++ TA+E A+ L + V+I PGYGMAVA+ + +A+I +
Sbjct 289 GEHREITAEETAELLKNSHSVIITPGYGMAVAQAQYPVAEITE 331
> dre:406619 nnt, wu:fa20d10, wu:fc86a04, zgc:76979; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P)
transhydrogenase [EC:1.6.1.2]
Length=1079
Score = 34.7 bits (78), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 16 GAVQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAK 58
G + D+ D + A ++IVPGYG+ A+ + +AD+ K
Sbjct 903 GTHTEVNVDQTVDLIKEAHNIIIVPGYGLCAAKAQYPIADLVK 945
> hsa:23530 NNT, MGC126502, MGC126503; nicotinamide nucleotide
transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 13/44 (29%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 16 GAVQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAKV 59
G + D D + A ++I PGYG+ A+ + +AD+ K+
Sbjct 907 GTHTEINLDNAIDMIREANSIIITPGYGLCAAKAQYPIADLVKM 950
> xla:447528 nnt, MGC83563; nicotinamide nucleotide transhydrogenase
(EC:1.6.1.2); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1086
Score = 32.0 bits (71), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 12/44 (27%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 16 GAVQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAKV 59
G + D + + A ++I PGYG+ A+ + +AD+ K+
Sbjct 907 GTHTEINLDNAVEYIREANNIIITPGYGLCAAKAQYPIADLVKI 950
> mmu:18115 Nnt, 4930423F13Rik, AI323702, BB168308; nicotinamide
nucleotide transhydrogenase (EC:1.6.1.2); K00323 NAD(P) transhydrogenase
[EC:1.6.1.2]
Length=1086
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 12/45 (26%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 15 QGAVQQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAKV 59
G + D + + A ++I PGYG+ A+ + +AD+ K+
Sbjct 906 SGTHTEINLDNAVEMIREANSIVITPGYGLCAAKAQYPIADLVKM 950
> cel:F55F10.1 hypothetical protein; K14572 midasin
Length=4368
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 23/41 (56%), Gaps = 3/41 (7%)
Query 19 QQTTADEVADELLAARKVLIVPGYGMAVARCRSELADIAKV 59
++ T DE D+ L ++L+ GY + RCR+E D+ V
Sbjct 923 KRMTTDETTDDWL---QILVNHGYFLLAGRCRNEKDDVTVV 960
Lambda K H
0.321 0.131 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2038271796
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40