bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3054_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
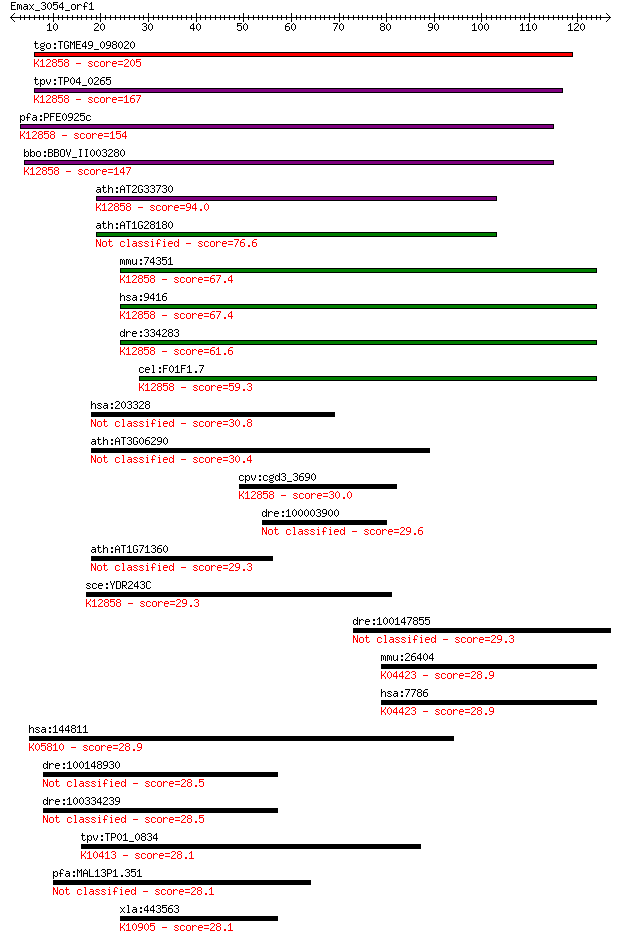
tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putativ... 205 3e-53
tpv:TP04_0265 small nuclear ribonucleoprotein; K12858 ATP-depe... 167 9e-42
pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA... 154 5e-38
bbo:BBOV_II003280 18.m06276; DEAD box RNA helicase; K12858 ATP... 147 6e-36
ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-depe... 94.0 1e-19
ath:AT1G28180 ATP binding / ATP-dependent helicase/ helicase/ ... 76.6 2e-14
mmu:74351 Ddx23, 3110082M05Rik, 4921506D17Rik; DEAD (Asp-Glu-A... 67.4 1e-11
hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEA... 67.4 1e-11
dre:334283 ddx23, wu:fi39b12, zgc:63742; DEAD (Asp-Glu-Ala-Asp... 61.6 6e-10
cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member (d... 59.3 3e-09
hsa:203328 SUSD3, MGC26847; sushi domain containing 3 30.8
ath:AT3G06290 hypothetical protein 30.4 1.4
cpv:cgd3_3690 U5 snRNP 100 kD protein ; K12858 ATP-dependent R... 30.0 1.9
dre:100003900 zgc:173927 29.6 2.7
ath:AT1G71360 hypothetical protein 29.3 3.0
sce:YDR243C PRP28; Prp28p (EC:3.6.1.-); K12858 ATP-dependent R... 29.3 3.3
dre:100147855 hypothetical LOC100147855 29.3 3.6
mmu:26404 Map3k12, DLK, MUK, Zpk; mitogen-activated protein ki... 28.9 4.3
hsa:7786 MAP3K12, DLK, MEKK12, MUK, ZPK, ZPKP1; mitogen-activa... 28.9 4.4
hsa:144811 C13orf31, DKFZp686D11119, FLJ38725; chromosome 13 o... 28.9 4.6
dre:100148930 hypothetical LOC100148930 28.5 6.1
dre:100334239 hypothetical protein LOC100334239 28.5 6.1
tpv:TP01_0834 hypothetical protein; K10413 dynein heavy chain ... 28.1 6.4
pfa:MAL13P1.351 conserved Plasmodium protein, unknown function 28.1 6.8
xla:443563 rad26, ATRIP; RAD26 protein; K10905 ATR interacting... 28.1 7.0
> tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putative
(EC:2.7.11.25); K12858 ATP-dependent RNA helicase DDX23/PRP28
[EC:3.6.4.13]
Length=1158
Score = 205 bits (521), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 96/113 (84%), Positives = 106/113 (93%), Gaps = 0/113 (0%)
Query 6 SLLADLKLLNLPEQELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDAS 65
S LADL+LLNLPEQELRA+QQE+ELEQIR HYLGM+ EKKKIQKPSEKFRNIFNFEW+ +
Sbjct 548 SSLADLRLLNLPEQELRARQQERELEQIRNHYLGMRTEKKKIQKPSEKFRNIFNFEWNDA 607
Query 66 EDTMRGDNNPLYQNRIEPQLLFGRGFRAGMDIREQRKANNFYDELVKRRQEYE 118
EDT +GDNNPLYQ R+EPQLLFGRGFRAGMDIREQRK NNFYDELVKRRQE++
Sbjct 608 EDTCKGDNNPLYQERMEPQLLFGRGFRAGMDIREQRKQNNFYDELVKRRQEHQ 660
> tpv:TP04_0265 small nuclear ribonucleoprotein; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=744
Score = 167 bits (422), Expect = 9e-42, Method: Compositional matrix adjust.
Identities = 79/111 (71%), Positives = 93/111 (83%), Gaps = 0/111 (0%)
Query 6 SLLADLKLLNLPEQELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDAS 65
S LA L LL LPE E+R + EKELEQIR+HYLG+ KEKKK+ KPSEKF+ IFNFEWD S
Sbjct 159 SDLAHLNLLKLPESEVREELIEKELEQIRLHYLGLNKEKKKVLKPSEKFKTIFNFEWDES 218
Query 66 EDTMRGDNNPLYQNRIEPQLLFGRGFRAGMDIREQRKANNFYDELVKRRQE 116
EDT + +NNP+YQ+R EPQLLFGRGFRAG+D+REQRK NNFYDEL ++R E
Sbjct 219 EDTTKFENNPIYQDRPEPQLLFGRGFRAGIDVREQRKKNNFYDELSRKRAE 269
> pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA
helicase DDX23/PRP28 [EC:3.6.4.13]
Length=1123
Score = 154 bits (390), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 77/112 (68%), Positives = 90/112 (80%), Gaps = 1/112 (0%)
Query 3 KEASLLADLKLLNLPEQELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEW 62
K S LA+L +LNL + R +EKELE I+ YLG+ K KKKIQKPSEKFRNIFNFEW
Sbjct 427 KPESSLAELNMLNLSNIQ-RDNLKEKELEIIKQQYLGLNKTKKKIQKPSEKFRNIFNFEW 485
Query 63 DASEDTMRGDNNPLYQNRIEPQLLFGRGFRAGMDIREQRKANNFYDELVKRR 114
D SEDT R D+NPLYQNR+EPQLLFGRG+ AG+D+REQRK NNFYD+LV+ R
Sbjct 486 DQSEDTSRNDSNPLYQNRLEPQLLFGRGYIAGIDVREQRKKNNFYDKLVQNR 537
> bbo:BBOV_II003280 18.m06276; DEAD box RNA helicase; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=714
Score = 147 bits (372), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 73/111 (65%), Positives = 86/111 (77%), Gaps = 0/111 (0%)
Query 4 EASLLADLKLLNLPEQELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWD 63
E S LA KLL LP E+R + EKELEQIR HYLG K K+K++K +EKFRN+F+FEWD
Sbjct 122 ENSDLAKKKLLTLPNSEVRPRMVEKELEQIREHYLGNKPTKQKVRKLTEKFRNVFHFEWD 181
Query 64 ASEDTMRGDNNPLYQNRIEPQLLFGRGFRAGMDIREQRKANNFYDELVKRR 114
S+DT R DNNP+YQNR EPQLLFGRG RAGMD +EQRK +FYD+L K R
Sbjct 182 NSDDTSRNDNNPIYQNRPEPQLLFGRGCRAGMDPKEQRKHADFYDKLSKLR 232
> ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=733
Score = 94.0 bits (232), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 50/84 (59%), Positives = 65/84 (77%), Gaps = 3/84 (3%)
Query 19 QELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDASEDTMRGDNNPLYQ 78
++L +++EKEL+ I+ YLG KK KK++ +PSEKFR F+F+W+ +EDT R D N LYQ
Sbjct 149 EKLVEREKEKELDAIKEQYLGGKKPKKRVIRPSEKFR--FSFDWENTEDTSR-DMNVLYQ 205
Query 79 NRIEPQLLFGRGFRAGMDIREQRK 102
N E QLLFGRGFRAGMD REQ+K
Sbjct 206 NPHEAQLLFGRGFRAGMDRREQKK 229
> ath:AT1G28180 ATP binding / ATP-dependent helicase/ helicase/
nucleic acid binding
Length=614
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 42/85 (49%), Positives = 58/85 (68%), Gaps = 5/85 (5%)
Query 19 QELRAKQQEKELEQIRMHYLGMKKEKKK-IQKPSEKFRNIFNFEWDASEDTMRGDNNPLY 77
++L ++EKE+ ++ YLG K KK+ I KPS+ FR F+W+ +EDT+ G+ N LY
Sbjct 95 EKLEMVKREKEINAMKEQYLGTTKPKKRVIMKPSKNFR----FDWENTEDTLSGEMNVLY 150
Query 78 QNRIEPQLLFGRGFRAGMDIREQRK 102
QN E Q LFGRG RAG+D REQ+K
Sbjct 151 QNPHEAQPLFGRGCRAGIDRREQKK 175
> mmu:74351 Ddx23, 3110082M05Rik, 4921506D17Rik; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 23 (EC:3.6.1.-); K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=819
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/101 (42%), Positives = 69/101 (68%), Gaps = 6/101 (5%)
Query 24 KQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDASEDTMRGDNNPLYQNRIEP 83
K + KEL I+ YLG K++++ + +++ F FEWDASEDT D NPLY+ R +
Sbjct 240 KDKSKELHAIKERYLGGIKKRRRTRHLNDRK---FVFEWDASEDTSI-DYNPLYKERHQV 295
Query 84 QLLFGRGFRAGMDIREQ-RKANNFYDELVKRRQEYELKQRQ 123
QLL GRGF AG+D+++Q R+ + FY +L+++R+ E K+++
Sbjct 296 QLL-GRGFIAGIDLKQQKREQSRFYGDLMEKRRTLEEKEQE 335
> hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 23 (EC:3.6.4.13); K12858
ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=820
Score = 67.4 bits (163), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 43/101 (42%), Positives = 69/101 (68%), Gaps = 6/101 (5%)
Query 24 KQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDASEDTMRGDNNPLYQNRIEP 83
K + KEL I+ YLG K++++ + +++ F FEWDASEDT D NPLY+ R +
Sbjct 241 KDKSKELHAIKERYLGGIKKRRRTRHLNDRK---FVFEWDASEDTSI-DYNPLYKERHQV 296
Query 84 QLLFGRGFRAGMDIREQ-RKANNFYDELVKRRQEYELKQRQ 123
QLL GRGF AG+D+++Q R+ + FY +L+++R+ E K+++
Sbjct 297 QLL-GRGFIAGIDLKQQKREQSRFYGDLMEKRRTLEEKEQE 336
> dre:334283 ddx23, wu:fi39b12, zgc:63742; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 23 (EC:3.6.1.-); K12858 ATP-dependent RNA
helicase DDX23/PRP28 [EC:3.6.4.13]
Length=807
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 39/101 (38%), Positives = 67/101 (66%), Gaps = 6/101 (5%)
Query 24 KQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDASEDTMRGDNNPLYQNRIEP 83
K + KEL I+ YLG K++++ + +++ F FEWDASEDT D NP+Y+ + +
Sbjct 228 KDKGKELVAIKERYLGGMKKRRRTRHLNDRK---FVFEWDASEDTSI-DYNPIYKEKHQV 283
Query 84 QLLFGRGFRAGMDIREQ-RKANNFYDELVKRRQEYELKQRQ 123
L +GRGF AG+D+++Q R + FY +L+++R+ E K+++
Sbjct 284 HL-YGRGFIAGIDLKQQKRDQSRFYGDLMEKRRTNEEKEQE 323
> cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member
(ddx-23); K12858 ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=730
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 40/97 (41%), Positives = 59/97 (60%), Gaps = 5/97 (5%)
Query 28 KELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDASEDTMRGDNNPLYQNRIEPQLLF 87
K + ++ YLG +KEKKK + F F+WDA EDT + D N LYQ+R E Q F
Sbjct 154 KMADAVKDRYLGKQKEKKK--RGRRLHEKKFVFDWDAGEDTSQ-DYNKLYQSRHEIQ-FF 209
Query 88 GRGFRAGMDIREQRK-ANNFYDELVKRRQEYELKQRQ 123
GRG AG D+ Q+K N+FY E+++ R+ + K+++
Sbjct 210 GRGSVAGTDVNAQKKEKNSFYQEMMENRRTVDEKEQE 246
> hsa:203328 SUSD3, MGC26847; sushi domain containing 3
Length=255
Score = 30.8 bits (68), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 28/51 (54%), Gaps = 1/51 (1%)
Query 18 EQELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDASEDT 68
+L ++ ++++LE ++ YLG+K K + PS+ N +F D E T
Sbjct 143 SAQLWSQLKDEDLETVQAAYLGLKHFNKPVSGPSQAHDN-HSFTTDHGEST 192
> ath:AT3G06290 hypothetical protein
Length=1697
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 39/76 (51%), Gaps = 6/76 (7%)
Query 18 EQELRAKQQEK---ELEQIRMHYLGMKKEKKKIQKPSEKFRN--IFNFEWDASEDTMRGD 72
E++ RAK+ + ELE I + ++ K + K + N FN ++S D ++GD
Sbjct 409 EEQARAKRLARFKGELEPIADRPVDIQLTKSPVNKTMKPLDNKQTFN-SLESSRDALKGD 467
Query 73 NNPLYQNRIEPQLLFG 88
P Y+N +P L+ G
Sbjct 468 ALPDYENSEQPSLIIG 483
> cpv:cgd3_3690 U5 snRNP 100 kD protein ; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=529
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 49 KPSEKFRNIFNFEWDASEDTMRGDNNPLYQNRI 81
K ++ R + FEW +EDT +N LY NR+
Sbjct 46 KSKKRHRGMLQFEWSNNEDTFSLGSNELYMNRL 78
> dre:100003900 zgc:173927
Length=481
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 10/26 (38%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 54 FRNIFNFEWDASEDTMRGDNNPLYQN 79
F NI +++ S+D + G N+PLY++
Sbjct 206 FINIMTYDFHGSQDNITGHNSPLYRD 231
> ath:AT1G71360 hypothetical protein
Length=596
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 18 EQELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFR 55
E +L A ++EKE+E +R+ GMK+ ++ +K + + R
Sbjct 470 EMDLEASKREKEVETMRLEVEGMKEREENTKKEAMEMR 507
> sce:YDR243C PRP28; Prp28p (EC:3.6.1.-); K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=588
Score = 29.3 bits (64), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 33/64 (51%), Gaps = 3/64 (4%)
Query 17 PEQELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFNFEWDASEDTMRGDNNPL 76
P Q AK + K++ + +KK+ PS++ + F+F W+ SEDT+ G +P+
Sbjct 54 PTQSDSAKVEIKKVNSRDDSFFNETNDKKR--NPSKQNGSKFHFSWNESEDTLSG-YDPI 110
Query 77 YQNR 80
R
Sbjct 111 VSTR 114
> dre:100147855 hypothetical LOC100147855
Length=323
Score = 29.3 bits (64), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 4/58 (6%)
Query 73 NNPLYQNRIEP-QLLFGRGFRAGMDIREQRKANNFYDELVKRRQEYELKQRQ---RGA 126
N+PL ++ P QLL GR R + Q + +D + +E +LKQ+Q RGA
Sbjct 161 NSPLDGVKLSPAQLLMGRRLRTRLPTSAQLQKPQLFDNVHHSIKERQLKQKQYFDRGA 218
> mmu:26404 Map3k12, DLK, MUK, Zpk; mitogen-activated protein
kinase kinase kinase 12 (EC:2.7.11.25); K04423 mitogen-activated
protein kinase kinase kinase 12 [EC:2.7.11.25]
Length=888
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Query 79 NRIEPQLLFGR--GFRAGMDIREQ-----RKANNFYDELVKRRQEYELKQRQ 123
+R+E +L+ R R +DIRE +ANN Y EL + ELK+R+
Sbjct 438 HRLEEELVMRRREELRHALDIREHYERKLERANNLYMELNALMLQLELKERE 489
> hsa:7786 MAP3K12, DLK, MEKK12, MUK, ZPK, ZPKP1; mitogen-activated
protein kinase kinase kinase 12 (EC:2.7.11.25); K04423
mitogen-activated protein kinase kinase kinase 12 [EC:2.7.11.25]
Length=892
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Query 79 NRIEPQLLFGR--GFRAGMDIREQ-----RKANNFYDELVKRRQEYELKQRQ 123
+R+E +L+ R R +DIRE +ANN Y EL + ELK+R+
Sbjct 438 HRLEEELVMRRREELRHALDIREHYERKLERANNLYMELNALMLQLELKERE 489
> hsa:144811 C13orf31, DKFZp686D11119, FLJ38725; chromosome 13
open reading frame 31; K05810 conserved hypothetical protein
Length=430
Score = 28.9 bits (63), Expect = 4.6, Method: Composition-based stats.
Identities = 21/96 (21%), Positives = 48/96 (50%), Gaps = 13/96 (13%)
Query 5 ASLLADLKLLNLPEQE-----LRAKQQEKELEQIRMHYLGMKKEKKKIQKP--SEKFRNI 57
++LL + ++++ P ++ K EK L I++ + + +K + K + F ++
Sbjct 70 SALLEEFEIVSCPSMAATLYTIKQKIDEKNLSSIKVI---VPRHRKTLMKAFIDQLFTDV 126
Query 58 FNFEWDASEDTMRGDNNPLYQNRIEPQLLFGRGFRA 93
+NFE++ + T RG L++ IE ++ + R
Sbjct 127 YNFEFEDLQVTFRGG---LFKQSIEINVITAQELRG 159
> dre:100148930 hypothetical LOC100148930
Length=659
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 33/53 (62%), Gaps = 4/53 (7%)
Query 8 LADLKLLNLPEQELRAKQQEKELEQIRMHYLGMKKEK----KKIQKPSEKFRN 56
L + KL + E EL+ K+QEKE E ++ Y +++EK KK+Q+ EK ++
Sbjct 599 LTEKKLKQILEYELKLKEQEKEQESLKQEYKHLQQEKEDDIKKLQQEVEKLKS 651
> dre:100334239 hypothetical protein LOC100334239
Length=659
Score = 28.5 bits (62), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 33/53 (62%), Gaps = 4/53 (7%)
Query 8 LADLKLLNLPEQELRAKQQEKELEQIRMHYLGMKKEK----KKIQKPSEKFRN 56
L + KL + E EL+ K+QEKE E ++ Y +++EK KK+Q+ EK ++
Sbjct 599 LTEKKLKQILEYELKLKEQEKEQESLKQEYKHLQQEKEDDIKKLQQEVEKLKS 651
> tpv:TP01_0834 hypothetical protein; K10413 dynein heavy chain
1, cytosolic
Length=1214
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 31/84 (36%), Gaps = 13/84 (15%)
Query 16 LPEQELRAKQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRNIFN-------------FEW 62
L EQ L + +KE+EQ + YL +E IQ E N F W
Sbjct 324 LAEQNLLISESQKEIEQNQSDYLKNIQESSTIQSEIEIIENKLILSKKVVDDLTNELFRW 383
Query 63 DASEDTMRGDNNPLYQNRIEPQLL 86
+ + DN L N I L+
Sbjct 384 NKRVSEIENDNKYLLSNSIMEALM 407
> pfa:MAL13P1.351 conserved Plasmodium protein, unknown function
Length=2868
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 10 DLKLLNLPEQELRAKQQEKELEQIRM-HYLGMKKEKKKIQKPSEKFRNIFNFEWD 63
DLK+ + E+ + +EKE+ +M Y + K+K I+K ++N+ N E D
Sbjct 1884 DLKIEENRKDEVTIEYKEKEIYFSKMLRYNNLVKQKNAIEKLVSHYKNVLNIEED 1938
> xla:443563 rad26, ATRIP; RAD26 protein; K10905 ATR interacting
protein
Length=798
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 24 KQQEKELEQIRMHYLGMKKEKKKIQKPSEKFRN 56
+Q E LEQ ++ ++ M+KEK +IQ E+ N
Sbjct 179 RQTESNLEQQKISHVLMEKEKSQIQSEKERELN 211
Lambda K H
0.317 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2059772308
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40