bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3076_orf2
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
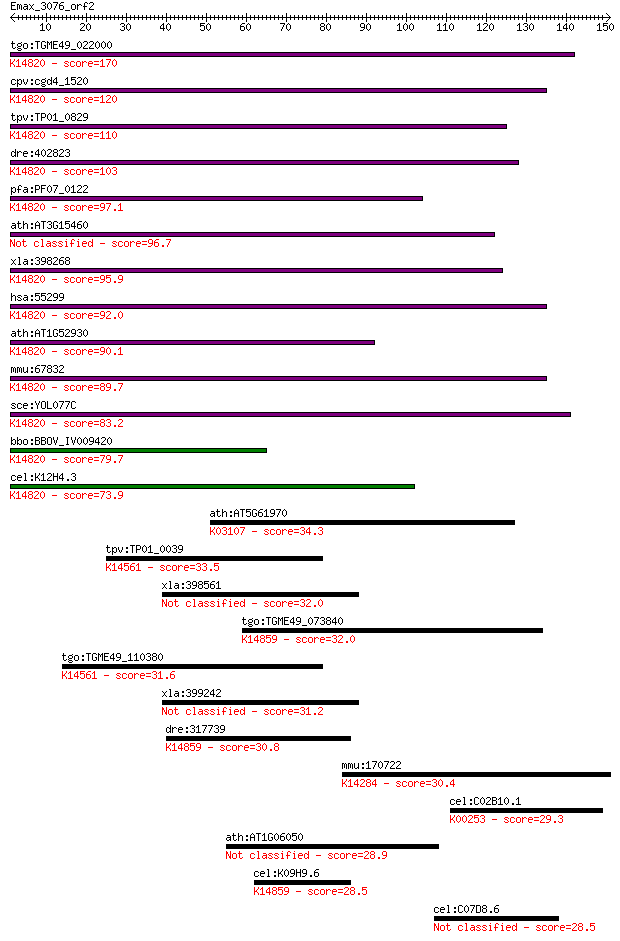
tgo:TGME49_022000 ribosome biogenesis protein Brix, putative ;... 170 2e-42
cpv:cgd4_1520 Brx1p nucleolar protein required for biogenesis ... 120 1e-27
tpv:TP01_0829 hypothetical protein; K14820 ribosome biogenesis... 110 2e-24
dre:402823 bxdc2; brix domain containing 2; K14820 ribosome bi... 103 3e-22
pfa:PF07_0122 nucleolus BRIX protein, putative; K14820 ribosom... 97.1 2e-20
ath:AT3G15460 brix domain-containing protein 96.7 3e-20
xla:398268 brix1, brix, bxdc2; BRX1, biogenesis of ribosomes, ... 95.9 4e-20
hsa:55299 BRIX1, BRIX, BXDC2, FLJ11100; BRX1, biogenesis of ri... 92.0 7e-19
ath:AT1G52930 brix domain-containing protein; K14820 ribosome ... 90.1 2e-18
mmu:67832 Brix1, 1110064N10Rik, Bxdc2, C76935; BRX1, biogenesi... 89.7 3e-18
sce:YOL077C BRX1; Brx1p; K14820 ribosome biogenesis protein BRX1 83.2 3e-16
bbo:BBOV_IV009420 23.m05729; ribosome biogenesis Brix protein;... 79.7 3e-15
cel:K12H4.3 hypothetical protein; K14820 ribosome biogenesis p... 73.9 2e-13
ath:AT5G61970 signal recognition particle-related / SRP-relate... 34.3 0.17
tpv:TP01_0039 U3 small nuclear ribonucleoprotein; K14561 U3 sm... 33.5 0.27
xla:398561 ppan-a; peter pan homolog 32.0 0.73
tgo:TGME49_073840 ribosome biogenesis protein SSF1, putative ;... 32.0 0.73
tgo:TGME49_110380 U3 small nucleolar ribonucleoprotein protein... 31.6 0.97
xla:399242 ppan-b, MGC81614, ppan; peter pan homolog 31.2 1.2
dre:317739 ppan, cb642; peter pan homolog (Drosophila); K14859... 30.8 1.6
mmu:170722 Nxf7, Nx7, Nxf6, nfx-a1, nxf-a, nxf-a2; nuclear RNA... 30.4 2.3
cel:C02B10.1 ivd-1; IsoValeryl-CoA Dehydrogenase family member... 29.3 4.9
ath:AT1G06050 hypothetical protein 28.9 5.7
cel:K09H9.6 lpd-6; LiPid Depleted family member (lpd-6); K1485... 28.5 7.8
cel:C07D8.6 hypothetical protein 28.5 7.9
> tgo:TGME49_022000 ribosome biogenesis protein Brix, putative
; K14820 ribosome biogenesis protein BRX1
Length=517
Score = 170 bits (430), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 85/143 (59%), Positives = 108/143 (75%), Gaps = 5/143 (3%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTD-SRIWFRHYQIAPLVMGEGGDANDPHRQT 59
KE+F+QVFGTPRNHPK+KPFFDHA++F+ D +RIWFRHYQIAPL+ GEGGDA+ P RQT
Sbjct 358 KEVFVQVFGTPRNHPKAKPFFDHALAFYKFDGNRIWFRHYQIAPLIGGEGGDADTPERQT 417
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRR 119
FIEIGPR VL+ VKIL+GSF GKT+W N ++ +RDL+ +R+ +A+R KEKR
Sbjct 418 FIEIGPRCVLEIVKILDGSFSGKTIWSNRNYICSRDLVALQRMGRAQSYAQRVQAKEKRT 477
Query 120 LYLESM-MEGEPRNKLAKEVVFG 141
L+ + +E P LA E VFG
Sbjct 478 ERLDKLHIEESP---LAMENVFG 497
> cpv:cgd4_1520 Brx1p nucleolar protein required for biogenesis
of the 60S ribosomal subunit ; K14820 ribosome biogenesis
protein BRX1
Length=353
Score = 120 bits (302), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 65/143 (45%), Positives = 87/143 (60%), Gaps = 18/143 (12%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
K+LF+QVFGTPR HPKSKPF DH +SF D +IWFRHYQIAP D N+ +Q
Sbjct 206 KDLFIQVFGTPRYHPKSKPFHDHVISFNFLDGKIWFRHYQIAPTT---ERDHNNVDKQVL 262
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVL------DFAKRQGQ 114
IEIGPRFVL+P+ IL+G+F G+ L+RN ++ P LR ++ + +R
Sbjct 263 IEIGPRFVLEPILILDGTFSGQVLYRNPDY------KSPTLLRNIIKQGYSSSYLQRVQS 316
Query 115 KEKRRLY---LESMMEGEPRNKL 134
K+KR Y L+S + P K+
Sbjct 317 KQKRSDYKDDLDSSLMNVPEKKM 339
> tpv:TP01_0829 hypothetical protein; K14820 ribosome biogenesis
protein BRX1
Length=294
Score = 110 bits (275), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 57/124 (45%), Positives = 75/124 (60%), Gaps = 5/124 (4%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
++L Q+FGTP HPKS PF DH +SFF D+RI RHYQI P N P Q
Sbjct 170 QKLISQIFGTPNQHPKSMPFHDHCLSFFYLDNRIHLRHYQITP---KNEFSVNSPDNQLL 226
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRRL 120
+EIGP+FVL+P+ IL+GSF G TLWRN +++ LM R R++ +R K+K L
Sbjct 227 VEIGPQFVLEPILILDGSFSGITLWRNSKYISP--LMVKRYERDMKLKKRRISNKKKVIL 284
Query 121 YLES 124
+ S
Sbjct 285 TIIS 288
> dre:402823 bxdc2; brix domain containing 2; K14820 ribosome
biogenesis protein BRX1
Length=383
Score = 103 bits (256), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 58/130 (44%), Positives = 76/130 (58%), Gaps = 21/130 (16%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KELF QVF TP +HPKS+PF DH +SF + D RIWFR+YQI DA +
Sbjct 218 KELFTQVFSTPLHHPKSQPFVDHVISFSIADHRIWFRNYQIV------EEDA------SL 265
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEK--- 117
+EIGPRFVL+P++I +GSF G TL+ N F P R +L A QKE+
Sbjct 266 VEIGPRFVLNPIRIFQGSFGGPTLYENPHF------QSPNAHRRMLRLAAAARQKERQMV 319
Query 118 RRLYLESMME 127
+++ +E ME
Sbjct 320 KQVRMEKRME 329
> pfa:PF07_0122 nucleolus BRIX protein, putative; K14820 ribosome
biogenesis protein BRX1
Length=416
Score = 97.1 bits (240), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 42/103 (40%), Positives = 68/103 (66%), Gaps = 6/103 (5%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KE+F+ VFG P HP SKPF+DH +FF + I+FRHYQI P+ + D+N+ ++Q
Sbjct 285 KEMFIHVFGVPNYHPLSKPFYDHCYNFFYVNDLIYFRHYQILPVTL---ADSNNVNKQQL 341
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLR 103
+EIGP+F L +KI E F G+ L+ N ++ ++ + P++++
Sbjct 342 VEIGPQFTLHIIKIFEECFKGRILYENEKY---QNYVTPQQVK 381
> ath:AT3G15460 brix domain-containing protein
Length=315
Score = 96.7 bits (239), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 48/122 (39%), Positives = 69/122 (56%), Gaps = 1/122 (0%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIA-PLVMGEGGDANDPHRQT 59
KE+ Q+FG P H KSKP+ DH F + D IWFR+YQI+ P + D + T
Sbjct 176 KEMLTQIFGIPEGHRKSKPYHDHVFVFSIVDDHIWFRNYQISVPHNESDKIARGDLDKMT 235
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRR 119
IE+GPRF L+P+KI GSF G TL+ N +V + + + FAK+ K +R+
Sbjct 236 LIEVGPRFCLNPIKIFGGSFGGTTLYENPFYVSPNQIRALEKRNKAGKFAKKIKAKTRRK 295
Query 120 LY 121
++
Sbjct 296 MH 297
> xla:398268 brix1, brix, bxdc2; BRX1, biogenesis of ribosomes,
homolog; K14820 ribosome biogenesis protein BRX1
Length=339
Score = 95.9 bits (237), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 53/123 (43%), Positives = 70/123 (56%), Gaps = 12/123 (9%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KEL Q+FGTPR HP+S+PF DH +F + D+RIWFR+YQI DA
Sbjct 175 KELLTQIFGTPRYHPRSQPFVDHIFTFSIADNRIWFRNYQII------EEDA------AL 222
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRRL 120
+EIGPRFVL+ +KI +GSF G TL+ N + R RL ++Q KE ++L
Sbjct 223 VEIGPRFVLNLIKIFKGSFGGPTLYENPHYQSPNMHRRMIRLATAAKVKEKQQVKEVQKL 282
Query 121 YLE 123
E
Sbjct 283 KKE 285
> hsa:55299 BRIX1, BRIX, BXDC2, FLJ11100; BRX1, biogenesis of
ribosomes, homolog (S. cerevisiae); K14820 ribosome biogenesis
protein BRX1
Length=353
Score = 92.0 bits (227), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 53/134 (39%), Positives = 76/134 (56%), Gaps = 17/134 (12%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KEL +Q+F TPR HPKS+PF DH +F + D+RIWFR++QI DA
Sbjct 182 KELLIQIFSTPRYHPKSQPFVDHVFTFTILDNRIWFRNFQII------EEDA------AL 229
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRRL 120
+EIGPRFVL+ +KI +GSF G TL+ N + M R +R + R+ Q+ K
Sbjct 230 VEIGPRFVLNLIKIFQGSFGGPTLYENPHYQSPN--MHRRVIRSITAAKYREKQQVKD-- 285
Query 121 YLESMMEGEPRNKL 134
++ + + EP+ L
Sbjct 286 -VQKLRKKEPKTLL 298
> ath:AT1G52930 brix domain-containing protein; K14820 ribosome
biogenesis protein BRX1
Length=320
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 56/92 (60%), Gaps = 1/92 (1%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIA-PLVMGEGGDANDPHRQT 59
KE+ QVFG P+ H KSKP+ DH F + D IWFR+YQI+ P + + T
Sbjct 180 KEMLTQVFGIPKEHRKSKPYHDHVFVFSIVDEHIWFRNYQISVPHNESDKIAKGGLDKMT 239
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFV 91
IE+GPRF L+P+KI GSF G TL+ N +V
Sbjct 240 LIEVGPRFCLNPIKIFAGSFGGPTLYENPLYV 271
> mmu:67832 Brix1, 1110064N10Rik, Bxdc2, C76935; BRX1, biogenesis
of ribosomes, homolog (S. cerevisiae); K14820 ribosome biogenesis
protein BRX1
Length=353
Score = 89.7 bits (221), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 52/134 (38%), Positives = 75/134 (55%), Gaps = 17/134 (12%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KE +Q+F TPR HPKS+PF DH +F + D+RIWFR++QI DA
Sbjct 182 KEFLIQIFSTPRYHPKSQPFVDHVFTFTILDNRIWFRNFQII------EEDA------AL 229
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRRL 120
+EIGPRFVL+ +KI +GSF G TL+ N + M R +R + R+ Q+ K
Sbjct 230 VEIGPRFVLNLIKIFQGSFGGPTLYENPHYQSPN--MHRRVIRSITAAKYRERQQVKD-- 285
Query 121 YLESMMEGEPRNKL 134
++ + + EP+ L
Sbjct 286 -VQKLRKKEPKTIL 298
> sce:YOL077C BRX1; Brx1p; K14820 ribosome biogenesis protein
BRX1
Length=291
Score = 83.2 bits (204), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 49/145 (33%), Positives = 81/145 (55%), Gaps = 12/145 (8%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
KEL + F P N KSKPF DH MSF + D +IW R Y+I+ + + +
Sbjct 153 KELLVHNFCVPPNARKSKPFIDHVMSFSIVDDKIWVRTYEISHSTKNKEEYEDGEEDISL 212
Query 61 IEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRRLREVLDFAKRQGQ-----K 115
+EIGPRFV+ + ILEGSF G ++ N ++V + +++R + ++ + AK + + K
Sbjct 213 VEIGPRFVMTVILILEGSFGGPKIYENKQYV-SPNVVRAQIKQQAAEEAKSRAEAAVERK 271
Query 116 EKRRLYLESMMEGEPRNKLAKEVVF 140
KRR E+++ +P L+ + +F
Sbjct 272 IKRR---ENVLAADP---LSNDALF 290
> bbo:BBOV_IV009420 23.m05729; ribosome biogenesis Brix protein;
K14820 ribosome biogenesis protein BRX1
Length=281
Score = 79.7 bits (195), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 38/64 (59%), Positives = 45/64 (70%), Gaps = 3/64 (4%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTF 60
K L QVFGTP NHPKSKPF DH +SFF D RI+FRH+QI+P+ E G N P +Q+
Sbjct 215 KSLISQVFGTPNNHPKSKPFHDHCLSFFYFDGRIFFRHFQISPI--NEYG-INKPEQQSL 271
Query 61 IEIG 64
EIG
Sbjct 272 TEIG 275
> cel:K12H4.3 hypothetical protein; K14820 ribosome biogenesis
protein BRX1
Length=352
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/102 (37%), Positives = 55/102 (53%), Gaps = 13/102 (12%)
Query 1 KELFMQVFGTPRNHPKSKPFFDHAMSFFVTD-SRIWFRHYQIAPLVMGEGGDANDPHRQT 59
K + MQ GTP +HP+S+PF DH +F V + +IWFR++QI +
Sbjct 185 KAVLMQTLGTPHHHPRSQPFVDHVFNFSVGEGDKIWFRNFQIVDESL------------Q 232
Query 60 FIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDLMRPRR 101
E+GPRFVL+ V++ GSF G L+ N +V + R R
Sbjct 233 LQEVGPRFVLEMVRLFAGSFEGAVLYDNPNYVSPNVIRREHR 274
> ath:AT5G61970 signal recognition particle-related / SRP-related;
K03107 signal recognition particle subunit SRP68
Length=605
Score = 34.3 bits (77), Expect = 0.17, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 43/78 (55%), Gaps = 4/78 (5%)
Query 51 DANDPHRQTFIEIGPRFVLDPVKILEGSFCGKTLWRNGEFVRTRDL--MRPRRLREVLDF 108
+ +DP ++ +I PRF ++ +++++ S L R+G++ R R R RRL + L F
Sbjct 12 EIDDPKSESSDQILPRFSINVLQLMKSSQAQHGL-RHGDYARYRRYCSARLRRLYKSLKF 70
Query 109 AKRQGQKEKRRLYLESMM 126
+G K RR LES +
Sbjct 71 THGRG-KYTRRAILESTV 87
> tpv:TP01_0039 U3 small nuclear ribonucleoprotein; K14561 U3
small nucleolar ribonucleoprotein protein IMP4
Length=302
Score = 33.5 bits (75), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 26/54 (48%), Gaps = 5/54 (9%)
Query 25 MSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQTFIEIGPRFVLDPVKILEGS 78
MSF +I FRH+ V E + + EIGPRFVL P KI G+
Sbjct 229 MSFINEKDKIIFRHH-----VWTEKKIGAEDKKVALKEIGPRFVLKPFKIELGT 277
> xla:398561 ppan-a; peter pan homolog
Length=564
Score = 32.0 bits (71), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 25/52 (48%), Gaps = 3/52 (5%)
Query 39 YQIAPLVMGEGGDANDPHRQTFI---EIGPRFVLDPVKILEGSFCGKTLWRN 87
+ I L G N +Q+ + EIGPR L VKI EG GK L+ N
Sbjct 247 HNITELPQACAGRGNMKAQQSAVRLTEIGPRMTLQLVKIEEGLSDGKVLYHN 298
> tgo:TGME49_073840 ribosome biogenesis protein SSF1, putative
; K14859 ribosome biogenesis protein SSF1/2
Length=673
Score = 32.0 bits (71), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 42/76 (55%), Gaps = 3/76 (3%)
Query 59 TFIEIGPRFVLDPVKILEGSFCGKTLWRNGEFV-RTRDLMRPRRLREVLDFAKRQGQKEK 117
+E+GPR L VKI E G+ L+ + F+ +T + +R + +E A+RQ Q+E+
Sbjct 410 ALVELGPRMELQLVKIEEDVCAGRVLYHH--FISKTPEELRMLKAKEAAVKAERQRQREE 467
Query 118 RRLYLESMMEGEPRNK 133
+E M+ + R++
Sbjct 468 MERAVELQMKKKRRDR 483
> tgo:TGME49_110380 U3 small nucleolar ribonucleoprotein protein,
putative (EC:3.1.2.15); K14561 U3 small nucleolar ribonucleoprotein
protein IMP4
Length=314
Score = 31.6 bits (70), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 20/81 (24%), Positives = 29/81 (35%), Gaps = 16/81 (19%)
Query 14 HPKSKPFFDHAMSFFVTDSRIWFRHYQIAPLVMGEGGDANDPHRQT-------------- 59
+P + P +F I FRHY G +N P +
Sbjct 209 YPPANPIAQRVQAFVNVGDEIHFRHYTWTDNRKKTSGASNTPGSSSADAEEEESSEKKGH 268
Query 60 --FIEIGPRFVLDPVKILEGS 78
+E+GPRF L P +I G+
Sbjct 269 LELVEVGPRFTLKPYRIDLGT 289
> xla:399242 ppan-b, MGC81614, ppan; peter pan homolog
Length=600
Score = 31.2 bits (69), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 25/52 (48%), Gaps = 3/52 (5%)
Query 39 YQIAPLVMGEGGDANDPHRQTFI---EIGPRFVLDPVKILEGSFCGKTLWRN 87
+ I L G N +Q+ + EIGPR L VKI EG GK L+ N
Sbjct 247 HNITELPQACAGRGNMKAQQSAVRLTEIGPRMTLQLVKIEEGLSDGKVLFHN 298
> dre:317739 ppan, cb642; peter pan homolog (Drosophila); K14859
ribosome biogenesis protein SSF1/2
Length=522
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 40 QIAPLVMGEGGDANDPHRQTFIEIGPRFVLDPVKILEGSFCGKTLW 85
++ + G G A+ EIGPR L VKI+EG G L+
Sbjct 251 ELPQVYSGRGNMASQQSAVRLTEIGPRMTLQLVKIVEGMGEGSVLY 296
> mmu:170722 Nxf7, Nx7, Nxf6, nfx-a1, nxf-a, nxf-a2; nuclear RNA
export factor 7; K14284 nuclear RNA export factor 1/2
Length=620
Score = 30.4 bits (67), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 33/69 (47%), Gaps = 2/69 (2%)
Query 84 LWRNGEFVRTRDLMRPRRLREVLDFAKRQGQKEKRRLYLESMMEGEPRN--KLAKEVVFG 141
LW G + + R +R VL+ + R+L L ++M + R K K++ G
Sbjct 323 LWLEGNPLCSTFPDRSSYIRAVLECFPELSYLDGRKLLLPTVMNTQERKLMKPCKDIFMG 382
Query 142 SDVVKQQSH 150
S+V+K Q H
Sbjct 383 SEVIKNQVH 391
> cel:C02B10.1 ivd-1; IsoValeryl-CoA Dehydrogenase family member
(ivd-1); K00253 isovaleryl-CoA dehydrogenase [EC:1.3.99.10]
Length=419
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 111 RQGQKEKRRLYLESMMEGEPRNKLA-KEVVFGSDVVKQQ 148
R G +E+R+ YL ++ GE LA E GSDVV +
Sbjct 136 RNGSEEQRKKYLPKLISGEHMGALAMSEAQAGSDVVSMK 174
> ath:AT1G06050 hypothetical protein
Length=313
Score = 28.9 bits (63), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 28/57 (49%), Gaps = 4/57 (7%)
Query 55 PHRQTFIEIGPRFVLDPVKILEGSFCGKTL---WRNGEFVRTRDLMRP-RRLREVLD 107
P TF+ GP++ D VKI G F K L W G + L P R+R+V+D
Sbjct 39 PSPDTFMVRGPKYFSDKVKIPAGDFLLKPLGFDWIKGPKKLSEILSYPSSRIRKVID 95
> cel:K09H9.6 lpd-6; LiPid Depleted family member (lpd-6); K14859
ribosome biogenesis protein SSF1/2
Length=573
Score = 28.5 bits (62), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 13/24 (54%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 62 EIGPRFVLDPVKILEGSFCGKTLW 85
EIGPR L+ VKI EG G+ L+
Sbjct 307 EIGPRLTLELVKIEEGIDEGEVLY 330
> cel:C07D8.6 hypothetical protein
Length=317
Score = 28.5 bits (62), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 107 DFAKRQGQKEKRRLYLESMMEGEPRNKLAKE 137
D AK + K +RL+L+ M G P + A E
Sbjct 285 DIAKLEESKNSQRLFLQDFMTGHPEDAFAAE 315
Lambda K H
0.325 0.141 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3134054208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40