bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3084_orf2
Length=158
Score E
Sequences producing significant alignments: (Bits) Value
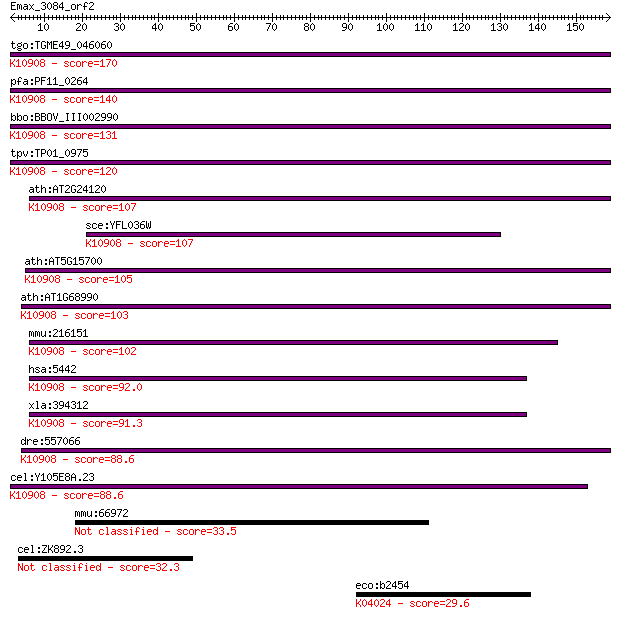
tgo:TGME49_046060 DNA-dependent RNA polymerase, putative (EC:2... 170 2e-42
pfa:PF11_0264 DNA-dependent RNA polymerase; K10908 DNA-directe... 140 2e-33
bbo:BBOV_III002990 17.m07284; DNA-dependent RNA polymerase; K1... 131 8e-31
tpv:TP01_0975 DNA-directed RNA polymerase; K10908 DNA-directed... 120 2e-27
ath:AT2G24120 SCA3; SCA3 (SCABRA 3); DNA binding / DNA-directe... 107 1e-23
sce:YFL036W RPO41; Mitochondrial RNA polymerase; single subuni... 107 2e-23
ath:AT5G15700 DNA-directed RNA polymerase (RPOT2); K10908 DNA-... 105 6e-23
ath:AT1G68990 DNA-directed RNA polymerase, mitochondrial (RPOM... 103 2e-22
mmu:216151 Polrmt, 1110018N15Rik, 4932416K13, MGC118526; polym... 102 6e-22
hsa:5442 POLRMT, APOLMT, MTRNAP, MTRPOL, h-mtRPOL; polymerase ... 92.0 8e-19
xla:394312 polrmt, mtRPO, polrmt-a; polymerase (RNA) mitochond... 91.3 1e-18
dre:557066 polrmt, si:dkeyp-74c5.5, wu:fb73g10; polymerase (RN... 88.6 7e-18
cel:Y105E8A.23 hypothetical protein; K10908 DNA-directed RNA p... 88.6 9e-18
mmu:66972 Slc25a23, 2310067G05Rik; solute carrier family 25 (m... 33.5 0.33
cel:ZK892.3 hypothetical protein 32.3 0.70
eco:b2454 eutJ, ECK2449, JW2438, yffW; predicted chaperonin, e... 29.6 4.8
> tgo:TGME49_046060 DNA-dependent RNA polymerase, putative (EC:2.7.7.6);
K10908 DNA-directed RNA polymerase, mitochondrial
[EC:2.7.7.6]
Length=2084
Score = 170 bits (431), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 86/183 (46%), Positives = 106/183 (57%), Gaps = 25/183 (13%)
Query 1 MRTKKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSA 60
MR KKWLD VS G+PVAW +P + EQPYRS + ++R+ Q H + V+GD
Sbjct 1887 MRLKKWLDEVSKTLNRLGLPVAWMLPLIHFGAEQPYRSGKAMEVRTVFQTHIVRVAGDDT 1946
Query 61 PVSRSKQRLGFPPNFIHSLDAAHMMLTALDC----------------IGT---------R 95
P S SKQRLGFPPNFIHSLDA HMMLTA C +G +
Sbjct 1947 PTSASKQRLGFPPNFIHSLDATHMMLTARRCLLRDPPPLVPPAFSQSVGGVETAWATPWK 2006
Query 96 HIDMAAVHDSYWAHAGCIDTLATALRQQFVALYQHNPLQSLYDSVKLRLPQQAAKLPPPP 155
+ AAVHDSYW HAG +D + T LRQ+FV LY L+ Y+ ++ RL + A LPPPP
Sbjct 2007 PMAFAAVHDSYWTHAGDVDRMRTILRQEFVRLYSQPILEEFYEGLRARLGRFAEDLPPPP 2066
Query 156 AKG 158
KG
Sbjct 2067 TKG 2069
> pfa:PF11_0264 DNA-dependent RNA polymerase; K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1531
Score = 140 bits (353), Expect = 2e-33, Method: Composition-based stats.
Identities = 64/158 (40%), Positives = 101/158 (63%), Gaps = 2/158 (1%)
Query 1 MRTKKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSA 60
M KKW + +S V +P+ W P +GL CEQPYR + + + LQ ++ +S ++
Sbjct 1361 MIIKKWFNNLSKVTNELNIPITWISP-IGLPCEQPYRLGNRILVNTPLQSISV-ISYKNS 1418
Query 61 PVSRSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATAL 120
+ ++KQRLGFPPNF+HSLDA+H+M+TA + + AAVHDSYWAHA +D + +
Sbjct 1419 QLHKNKQRLGFPPNFVHSLDASHLMMTAEKMVIQNNFSFAAVHDSYWAHACNVDIMNQFI 1478
Query 121 RQQFVALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
R+ F+ LY L+++Y++ ++RL + A K+P P +G
Sbjct 1479 RESFITLYNEPILENIYNNFRIRLGKFATKIPSSPEQG 1516
> bbo:BBOV_III002990 17.m07284; DNA-dependent RNA polymerase;
K10908 DNA-directed RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1088
Score = 131 bits (330), Expect = 8e-31, Method: Compositional matrix adjust.
Identities = 71/160 (44%), Positives = 99/160 (61%), Gaps = 8/160 (5%)
Query 1 MRTKKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSA 60
M KKWLD +S VPV W P +GL CEQPY + +R+ +Q +++S ++A
Sbjct 920 MGIKKWLDDISAAHNRLNVPVTWISP-IGLPCEQPYCKAVTKVVRTPIQ--AINIS-ENA 975
Query 61 P--VSRSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLAT 118
P V + KQ+L FPPNF+HSLDA+H+MLTA + + AAVHDSYW HA +D +
Sbjct 976 PSVVDKRKQKLAFPPNFVHSLDASHLMLTA-EVLFRDGFSFAAVHDSYWTHACDVDIMNK 1034
Query 119 ALRQQFVALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
A+R+QFV +Y L++LY S RL + +PPPP +G
Sbjct 1035 AIREQFVKMYNEPILENLYQSCNKRLRGEVT-IPPPPPRG 1073
> tpv:TP01_0975 DNA-directed RNA polymerase; K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1225
Score = 120 bits (301), Expect = 2e-27, Method: Composition-based stats.
Identities = 64/159 (40%), Positives = 90/159 (56%), Gaps = 9/159 (5%)
Query 1 MRTKKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSA 60
M K WLD +S V +PV W P +GL CEQPY + +R+ LQ ++VS +
Sbjct 1060 MSIKNWLDEISCVHNKFNIPVTWISP-IGLPCEQPYCKSVTKVVRTPLQ--AINVSKYVS 1116
Query 61 PVSRSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHI-DMAAVHDSYWAHAGCIDTLATA 119
V + +Q+L FPPNFIHSLDA+H+M+TA + +H A+VHDSYW H +D + T
Sbjct 1117 SVDKRRQKLAFPPNFIHSLDASHLMMTATEVF--KHTQSFASVHDSYWIHPDKVDMMHTY 1174
Query 120 LRQQFVALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
+R +F ++ L L+ SV L + PPP KG
Sbjct 1175 IRNEFFNMHTKPILDDLHKSV---LNKLQINFRPPPKKG 1210
> ath:AT2G24120 SCA3; SCA3 (SCABRA 3); DNA binding / DNA-directed
RNA polymerase; K10908 DNA-directed RNA polymerase, mitochondrial
[EC:2.7.7.6]
Length=993
Score = 107 bits (268), Expect = 1e-23, Method: Composition-based stats.
Identities = 61/153 (39%), Positives = 83/153 (54%), Gaps = 6/153 (3%)
Query 6 WLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSRS 65
WL + + + PV W P +GL QPY ++ IR++LQ L G++ V
Sbjct 832 WLGDCAKIIASDNHPVRWITP-LGLPVVQPYCRSERHLIRTSLQVLALQREGNTVDVR-- 888
Query 66 KQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQFV 125
KQR FPPNF+HSLD HMM+TA+ C ++ A VHDSYW HA +DT+ LR++FV
Sbjct 889 KQRTAFPPNFVHSLDGTHMMMTAVAC-REAGLNFAGVHDSYWTHACDVDTMNRILREKFV 947
Query 126 ALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
LY L+ L S + P PP P +G
Sbjct 948 ELYNTPILEDLLQSFQESYPNLV--FPPVPKRG 978
> sce:YFL036W RPO41; Mitochondrial RNA polymerase; single subunit
enzyme similar to those of T3 and T7 bacteriophages; requires
a specificity subunit encoded by MTF1 for promoter recognition
(EC:2.7.7.6); K10908 DNA-directed RNA polymerase, mitochondrial
[EC:2.7.7.6]
Length=1351
Score = 107 bits (267), Expect = 2e-23, Method: Composition-based stats.
Identities = 53/111 (47%), Positives = 75/111 (67%), Gaps = 6/111 (5%)
Query 21 VAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSA--PVSRSKQRLGFPPNFIHS 78
V W P +GL QPYR ++Q+ + LQ T+ +S A PV+ +Q+ G PPNFIHS
Sbjct 1108 VIWTTP-LGLPIVQPYREESKKQVETNLQ--TVFISDPFAVNPVNARRQKAGLPPNFIHS 1164
Query 79 LDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQFVALYQ 129
LDA+HM+L+A +C G + +D A+VHDSYW HA IDT+ LR+QF+ L++
Sbjct 1165 LDASHMLLSAAEC-GKQGLDFASVHDSYWTHASDIDTMNVVLREQFIKLHE 1214
> ath:AT5G15700 DNA-directed RNA polymerase (RPOT2); K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1011
Score = 105 bits (262), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 57/154 (37%), Positives = 82/154 (53%), Gaps = 6/154 (3%)
Query 5 KWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSR 64
+W + + + V W P +GL QPY + + ++++LQ +L D V R
Sbjct 849 RWFGECAKIIASENETVRWTTP-LGLPVVQPYHQMGTKLVKTSLQTLSLQHETDQVIVRR 907
Query 65 SKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQF 124
QR FPPNFIHSLD +HMM+TA+ C + A VHDS+W HA +D L LR++F
Sbjct 908 --QRTAFPPNFIHSLDGSHMMMTAVAC-KRAGVCFAGVHDSFWTHACDVDKLNIILREKF 964
Query 125 VALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
V LY L++L +S + P PP P +G
Sbjct 965 VELYSQPILENLLESFEQSFPH--LDFPPLPERG 996
> ath:AT1G68990 DNA-directed RNA polymerase, mitochondrial (RPOMT);
K10908 DNA-directed RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=976
Score = 103 bits (258), Expect = 2e-22, Method: Composition-based stats.
Identities = 57/155 (36%), Positives = 83/155 (53%), Gaps = 6/155 (3%)
Query 4 KKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVS 63
K W + + + V W P +GL QPYR + +++ LQ TL S ++ V
Sbjct 813 KSWFGDCAKIIASENNAVCWTTP-LGLPVVQPYRKPGRHLVKTTLQVLTL--SRETDKVM 869
Query 64 RSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQ 123
+Q F PNFIHSLD +HMM+TA+ C + A VHDS+W HA +D + T LR++
Sbjct 870 ARRQMTAFAPNFIHSLDGSHMMMTAVAC-NRAGLSFAGVHDSFWTHACDVDVMNTILREK 928
Query 124 FVALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
FV LY+ L++L +S + P PP P +G
Sbjct 929 FVELYEKPILENLLESFQKSFPD--ISFPPLPERG 961
> mmu:216151 Polrmt, 1110018N15Rik, 4932416K13, MGC118526; polymerase
(RNA) mitochondrial (DNA directed) (EC:2.7.7.6); K10908
DNA-directed RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1207
Score = 102 bits (253), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 57/140 (40%), Positives = 79/140 (56%), Gaps = 3/140 (2%)
Query 6 WLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVSRS 65
WL + + + AG PV W P +G+ QPY + Q++ LQ TL S D + +
Sbjct 1024 WLTESANLISHAGWPVEWVTP-LGIPIIQPYHRESKVQVKGGLQSITLTSSVDESQKPNT 1082
Query 66 -KQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQF 124
KQ+ GFPPNFIHSLD++HMMLTAL C + + +VHD +W HA I T+ R+QF
Sbjct 1083 LKQKNGFPPNFIHSLDSSHMMLTALHCY-RKGLIFVSVHDCFWTHAADIPTMNEVCREQF 1141
Query 125 VALYQHNPLQSLYDSVKLRL 144
V L+ L+ L +K R
Sbjct 1142 VRLHSQPILEDLAKFLKKRF 1161
> hsa:5442 POLRMT, APOLMT, MTRNAP, MTRPOL, h-mtRPOL; polymerase
(RNA) mitochondrial (DNA directed) (EC:2.7.7.6); K10908 DNA-directed
RNA polymerase, mitochondrial [EC:2.7.7.6]
Length=1230
Score = 92.0 bits (227), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 54/133 (40%), Positives = 74/133 (55%), Gaps = 4/133 (3%)
Query 6 WLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYR-SLQQQQIRSALQRHTLHVSGD-SAPVS 63
WL + + + G V W P +G+ QPYR + +QI +Q T +GD S +
Sbjct 1053 WLTESARLISHMGSVVEWVTP-LGVPVIQPYRLDSKVKQIGGGIQSITYTHNGDISRKPN 1111
Query 64 RSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQ 123
KQ+ GFPPNFIHSLD++HMMLTAL C + + +VHD YW HA + + R+Q
Sbjct 1112 TRKQKNGFPPNFIHSLDSSHMMLTALHCY-RKGLTFVSVHDCYWTHAADVSVMNQVCREQ 1170
Query 124 FVALYQHNPLQSL 136
FV L+ LQ L
Sbjct 1171 FVRLHSEPILQDL 1183
> xla:394312 polrmt, mtRPO, polrmt-a; polymerase (RNA) mitochondrial
(DNA directed); K10908 DNA-directed RNA polymerase, mitochondrial
[EC:2.7.7.6]
Length=1235
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 52/132 (39%), Positives = 71/132 (53%), Gaps = 3/132 (2%)
Query 6 WLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGD-SAPVSR 64
WL + + + +G V W+ P +GL QPY + S LQ L S D +
Sbjct 1053 WLTESARMISKSGNTVEWQTP-LGLPIIQPYHRAKPVLFHSNLQVVNLQSSHDVNERPDT 1111
Query 65 SKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQF 124
KQ+ FPPNFIHSLD+ HMMLTAL C + + +VHD +W H +D + R+QF
Sbjct 1112 MKQKNAFPPNFIHSLDSTHMMLTALHCY-SHGLTFVSVHDCFWTHPDTVDVMNKVCREQF 1170
Query 125 VALYQHNPLQSL 136
VAL+ LQ+L
Sbjct 1171 VALHSQPILQNL 1182
> dre:557066 polrmt, si:dkeyp-74c5.5, wu:fb73g10; polymerase (RNA)
mitochondrial (DNA directed); K10908 DNA-directed RNA polymerase,
mitochondrial [EC:2.7.7.6]
Length=1253
Score = 88.6 bits (218), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 55/155 (35%), Positives = 80/155 (51%), Gaps = 13/155 (8%)
Query 4 KKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSAPVS 63
++WL + + +G V W P +GL QPY ++ Q + + L DS
Sbjct 1097 QEWLTESARLIAKSGRTVEWVTP-LGLPIIQPYHRIRNQIVTCS--SCALFRKPDSM--- 1150
Query 64 RSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATALRQQ 123
KQ+ FPPNFIHSLD+ HMMLT+L C + +VHD YW HA +D + R+Q
Sbjct 1151 --KQKNAFPPNFIHSLDSTHMMLTSLHC-HRAGLTFVSVHDCYWTHAVTVDIMNRVCREQ 1207
Query 124 FVALYQHNPLQSLYDSVKLRLPQQAAKLPPPPAKG 158
FVAL+ Q + + + L Q+ ++ PP P G
Sbjct 1208 FVALHS----QPILEELSWFLLQKYSRRPPVPQTG 1238
> cel:Y105E8A.23 hypothetical protein; K10908 DNA-directed RNA
polymerase, mitochondrial [EC:2.7.7.6]
Length=1138
Score = 88.6 bits (218), Expect = 9e-18, Method: Composition-based stats.
Identities = 56/154 (36%), Positives = 77/154 (50%), Gaps = 16/154 (10%)
Query 1 MRTKKWLDAVSTVCTAAGVPVAWKVPAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDSA 60
M K W ++ + V W P +GL QPY L +++ + L A
Sbjct 966 MALKDWFRLIAKGSSDLMKTVEWITP-LGLPVVQPYCKLVERKGKLIL-----------A 1013
Query 61 PVSRSKQRLGFPPNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHAGCIDTLATAL 120
PV KQ FPPNF+HSLD+ HMMLT+L+C R I AAVHD +W HA +D +
Sbjct 1014 PVPM-KQVDAFPPNFVHSLDSTHMMLTSLNC-AQRGITFAAVHDCFWTHANSVDQMNEIC 1071
Query 121 RQQFVALYQHNPLQSLYDSVK--LRLPQQAAKLP 152
RQQFV+L+ ++ + K P+ A LP
Sbjct 1072 RQQFVSLHSQPIVEQCSEWFKKTYLTPKMAKILP 1105
> mmu:66972 Slc25a23, 2310067G05Rik; solute carrier family 25
(mitochondrial carrier; phosphate carrier), member 23; K14684
solute carrier family 25 (mitochondrial phosphate transporter),
member 23/24/25/41
Length=467
Score = 33.5 bits (75), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 45/98 (45%), Gaps = 9/98 (9%)
Query 18 GVPVAWKV-PAVGLLCEQPYRSLQQQQIRSALQRHTLHVSGDS----APVSRSKQRLGFP 72
GV W P GL E+ R LQ+++ R L H+L + D + + +S + LG
Sbjct 52 GVSSDWDADPDGGLSLEEFTRYLQEREQRLLLMFHSLDRNQDGHIDVSEIQQSFRALGIS 111
Query 73 PNFIHSLDAAHMMLTALDCIGTRHIDMAAVHDSYWAHA 110
SL+ A +L ++D GT ID D + H+
Sbjct 112 I----SLEQAEKILHSMDRDGTMTIDWQEWRDHFLLHS 145
> cel:ZK892.3 hypothetical protein
Length=541
Score = 32.3 bits (72), Expect = 0.70, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 24/49 (48%), Gaps = 4/49 (8%)
Query 3 TKKWLDAVSTVCTAAGV---PVAWKVPAVGLLCEQPYRSLQQQQIRSAL 48
T++W D S VC A P+ WK+P + EQ +R + + R L
Sbjct 213 TRRW-DTASYVCAAISALIFPILWKLPESPVFLEQKHRIEEANEAREQL 260
> eco:b2454 eutJ, ECK2449, JW2438, yffW; predicted chaperonin,
ethanolamine utilization protein; K04024 ethanolamine utilization
protein EutJ
Length=278
Score = 29.6 bits (65), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query 92 IGTRHIDMAAVHDSYWAHAGCID-TLATALRQQFVALYQHNPLQSLY 137
I RHI+ + D + A C+ +A R+QF AL H P SL+
Sbjct 212 IVARHIEGQGITDLWLAGGSCMQPGVAELFRKQFPALQVHLPQHSLF 258
Lambda K H
0.322 0.132 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3582056500
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40