bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3091_orf1
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
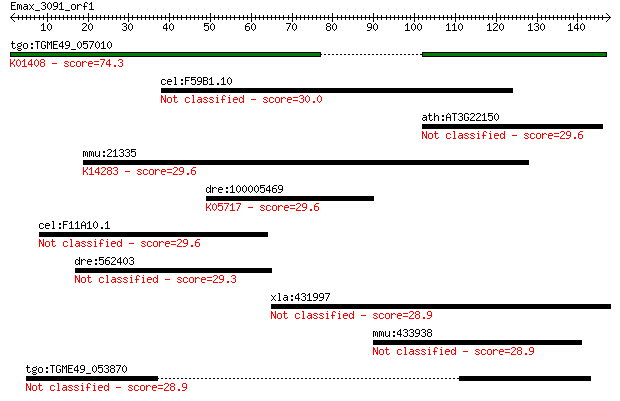
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 74.3 1e-13
cel:F59B1.10 hypothetical protein 30.0 2.5
ath:AT3G22150 pentatricopeptide (PPR) repeat-containing protein 29.6 3.2
mmu:21335 Tacc3, Aint, C86661, Eric1; transforming, acidic coi... 29.6 3.3
dre:100005469 fn1, Fn, fb80d10, fn2, wu:fb80d10; fibronectin 1... 29.6 3.6
cel:F11A10.1 lex-1; Lin-48 EXpression abnormal family member (... 29.6 3.9
dre:562403 hypothetical LOC562403 29.3 4.5
xla:431997 coro2a, MGC81182; coronin, actin binding protein, 2A 28.9 5.4
mmu:433938 Mn1, AA003644, AA009236; meningioma 1 28.9
tgo:TGME49_053870 hypothetical protein 28.9 5.9
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/76 (46%), Positives = 52/76 (68%), Gaps = 0/76 (0%)
Query 1 KEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAKLS 60
KE+A A+L K F E+F +S +I H +CF KRDLE+ YL+ +F+RKQL RT+ KL
Sbjct 868 KESARAELEKPTETFYEEFGRSWGQIANHGHCFNKRDLELIYLNTEFNRKQLSRTYTKLL 927
Query 61 DPSRRMVVKLIADLEP 76
+P+RR+V ++ + P
Sbjct 928 EPTRRLVGFVLNHVTP 943
Score = 46.2 bits (108), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 102 EMARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDMEV 146
E+ WKE+A A+L K F E+F +S +I H +CF KRD+E+
Sbjct 863 ELDHWKESARAELEKPTETFYEEFGRSWGQIANHGHCFNKRDLEL 907
> cel:F59B1.10 hypothetical protein
Length=428
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 44/87 (50%), Gaps = 3/87 (3%)
Query 38 LEVKYLDNDFSRKQLLRTFAKLSDPSRRMVVKLIADLEPEKEV-TLIGEAKAQDAALLRR 96
+EV N FS + LL + + PS + KLI + + LI + K Q+A+L+ +
Sbjct 45 MEVIGDGNGFSSRVLLIS-CNWTIPSAHLPKKLILKIVSFVHIQALIDKGKQQNASLITK 103
Query 97 GDTTEEMARWKEAAHAKLTKMEANFSE 123
+ E+M + E++ K+ E NF E
Sbjct 104 -EVEEQMYAYFESSCKKMHNQEMNFYE 129
> ath:AT3G22150 pentatricopeptide (PPR) repeat-containing protein
Length=820
Score = 29.6 bits (65), Expect = 3.2, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 19/44 (43%), Gaps = 0/44 (0%)
Query 102 EMARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDME 145
E +WK + E ++ +S EI + NCF RD E
Sbjct 739 EEQKWKSVDKVRRGMREKGLKKEVGRSGIEIAGYVNCFVSRDQE 782
> mmu:21335 Tacc3, Aint, C86661, Eric1; transforming, acidic coiled-coil
containing protein 3; K14283 transforming acidic
coiled-coil-containing protein 3
Length=630
Score = 29.6 bits (65), Expect = 3.3, Method: Composition-based stats.
Identities = 27/109 (24%), Positives = 50/109 (45%), Gaps = 14/109 (12%)
Query 19 FKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAKLSDPSRRMVVKLIADLEPEK 78
K S +++ A N + +LE+K D + K L K D ++ K + + E ++
Sbjct 426 LKYSQKDLDAVVNVMQQENLELKSKYEDLNTKYL--EMGKSVDEFEKIAYKSLEEAEKQR 483
Query 79 EVTLIGEAKAQDAALLRRGDTTEEMARWKEAAHAKLTKMEANFSEDFKK 127
E+ I E K Q ++ + ++ +A L ME +FS+ FK+
Sbjct 484 ELKEIAEDKIQ------------KVLKERDQLNADLNSMEKSFSDLFKR 520
> dre:100005469 fn1, Fn, fb80d10, fn2, wu:fb80d10; fibronectin
1; K05717 fibronectin 1
Length=2478
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Query 49 RKQLLRTFAKLSD--PSRRMVVKLIADLEPEKEVTLIGEAKAQ 89
R Q RTFA +S P ++K++A ++ LIG+A+ Q
Sbjct 2044 RPQAARTFASISGLIPGTEYIIKIVALNGAQRSTPLIGKARTQ 2086
> cel:F11A10.1 lex-1; Lin-48 EXpression abnormal family member
(lex-1)
Length=1291
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 8/60 (13%)
Query 8 LTKMEANFSEDFKKSAEEIFAHSNCFT----KRDLEVKYLDNDFSRKQLLRTFAKLSDPS 63
L+ ++ +F ED + EIF H+NC T +R + KY ++ + + T K+ DP+
Sbjct 825 LSTLDTSF-EDAPEYVTEIFRHANCITLNPSRRTIRQKYFEHVIEK---INTPPKVFDPT 880
> dre:562403 hypothetical LOC562403
Length=776
Score = 29.3 bits (64), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 30/48 (62%), Gaps = 2/48 (4%)
Query 17 EDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSRKQLLRTFAKLSDPSR 64
+D ++ AE +FAH +R+ + + + N + KQL +T +++SD SR
Sbjct 432 QDLRQHAE-VFAHQMLVKQRETQ-RQIQNAETEKQLQQTLSQVSDISR 477
> xla:431997 coro2a, MGC81182; coronin, actin binding protein,
2A
Length=525
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 38/83 (45%), Gaps = 2/83 (2%)
Query 65 RMVVKLIADLEPEKEVTLIGEAKAQDAALLRRGDTTEEMARWKEAAHAKLTKMEANFSED 124
+M K + ++E + E+ I E ++ A R DT + M+ + K E +
Sbjct 429 KMPTKTLWEIEYKTEI--IREDRSNKKAFERLQDTKQAMSNGFDIFECPPPKTENELLQM 486
Query 125 FKKSAEEIFAHSNCFTKRDMEVK 147
F K EEI + +RD+++K
Sbjct 487 FYKQQEEIRRLRDQINQRDVQIK 509
> mmu:433938 Mn1, AA003644, AA009236; meningioma 1
Length=1297
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 16/51 (31%), Positives = 24/51 (47%), Gaps = 0/51 (0%)
Query 90 DAALLRRGDTTEEMARWKEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFT 140
D AL+ GD + +A W++A EA+ K SA + +H C T
Sbjct 1211 DKALVDGGDEDKTLAPWEKAKSQNPNNKEAHDHPTNKASATQPGSHLQCLT 1261
> tgo:TGME49_053870 hypothetical protein
Length=4787
Score = 28.9 bits (63), Expect = 5.9, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 5 HAKLTKMEA--NFSEDFKKSAEEIFAHSNCFTKR 36
+ +L K+E FS DF KSA I H+ C KR
Sbjct 2244 YTRLGKLERLLRFSADFTKSAHHILLHAACLRKR 2277
Score = 28.9 bits (63), Expect = 5.9, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 111 HAKLTKMEA--NFSEDFKKSAEEIFAHSNCFTKR 142
+ +L K+E FS DF KSA I H+ C KR
Sbjct 2244 YTRLGKLERLLRFSADFTKSAHHILLHAACLRKR 2277
Lambda K H
0.315 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40