bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3097_orf1
Length=137
Score E
Sequences producing significant alignments: (Bits) Value
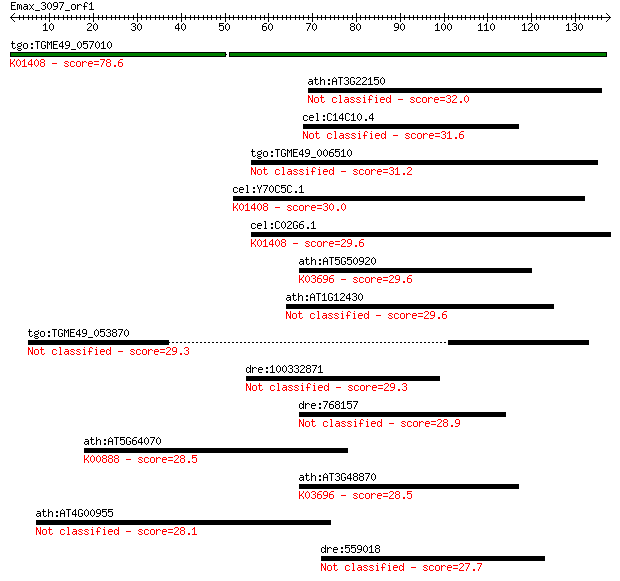
tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408 i... 78.6 6e-15
ath:AT3G22150 pentatricopeptide (PPR) repeat-containing protein 32.0 0.63
cel:C14C10.4 hypothetical protein 31.6 0.83
tgo:TGME49_006510 peptidase M16 domain containing protein (EC:... 31.2 0.91
cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24... 30.0 2.1
cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56] 29.6 2.7
ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptid... 29.6 2.8
ath:AT1G12430 ARK3; ARK3 (ARMADILLO REPEAT KINESIN 3); ATP bin... 29.6 2.9
tgo:TGME49_053870 hypothetical protein 29.3 3.5
dre:100332871 protein-kinase, interferon-inducible double stra... 29.3 4.2
dre:768157 cplx4a, CPLX4, si:ch211-233a1.5, zgc:153429; comple... 28.9 5.2
ath:AT5G64070 PI-4KBETA1 (PHOSPHATIDYLINOSITOL 4-OH KINASE BET... 28.5 6.5
ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding / n... 28.5 6.8
ath:AT4G00955 hypothetical protein 28.1 9.1
dre:559018 clul1, wu:fb51b10; clusterin-like 1 (retinal) 27.7 9.8
> tgo:TGME49_057010 insulysin, putative (EC:3.4.24.56); K01408
insulysin [EC:3.4.24.56]
Length=953
Score = 78.6 bits (192), Expect = 6e-15, Method: Composition-based stats.
Identities = 35/86 (40%), Positives = 57/86 (66%), Gaps = 0/86 (0%)
Query 51 ASTALLQCFVKGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEAN 110
+S L+C ++G++ HPDE+ +ID+EL K ++L ++ + E+ WKE+A A+L K
Sbjct 822 SSVESLRCVLEGSRKHPDEIADLIDKELWKMNDHLQSISDGELDHWKESARAELEKPTET 881
Query 111 FSEDFKKSAEEIFAHSNCFTKRDLEV 136
F E+F +S +I H +CF KRDLE+
Sbjct 882 FYEEFGRSWGQIANHGHCFNKRDLEL 907
Score = 47.0 bits (110), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/49 (44%), Positives = 33/49 (67%), Gaps = 0/49 (0%)
Query 1 KEAAHAKLTKMEANFSEDFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSQ 49
KE+A A+L K F E+F +S +I H +CF KRDLE+ YL+ +F++
Sbjct 868 KESARAELEKPTETFYEEFGRSWGQIANHGHCFNKRDLELIYLNTEFNR 916
> ath:AT3G22150 pentatricopeptide (PPR) repeat-containing protein
Length=820
Score = 32.0 bits (71), Expect = 0.63, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 34/76 (44%), Gaps = 10/76 (13%)
Query 69 EVVKMIDEELSK---AKEY------LANMPEAEMARWKEAAHAKLTKMEANFSEDFKKSA 119
E+ + + E L+K K + L+NM AE +WK + E ++ +S
Sbjct 708 ELAETVSERLAKFDKGKNFSGYEVLLSNM-YAEEQKWKSVDKVRRGMREKGLKKEVGRSG 766
Query 120 EEIFAHSNCFTKRDLE 135
EI + NCF RD E
Sbjct 767 IEIAGYVNCFVSRDQE 782
> cel:C14C10.4 hypothetical protein
Length=1058
Score = 31.6 bits (70), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 11/60 (18%)
Query 68 DEVVKMIDEELSKAKEYLANMPE-----------AEMARWKEAAHAKLTKMEANFSEDFK 116
D V++ +EEL+ A+ L + PE A M RW+ +L K +NFS D K
Sbjct 650 DPRVQLNNEELTDARNVLKSSPEKAELLGKLRKSATMQRWRATDVNELLKELSNFSNDVK 709
> tgo:TGME49_006510 peptidase M16 domain containing protein (EC:4.1.1.70
3.4.24.13 3.4.24.56 3.4.21.10 3.4.24.35 3.2.1.91)
Length=2435
Score = 31.2 bits (69), Expect = 0.91, Method: Composition-based stats.
Identities = 19/81 (23%), Positives = 42/81 (51%), Gaps = 2/81 (2%)
Query 56 LQCFVKGAKAHPDEVVKMIDEELSKAKEYLAN--MPEAEMARWKEAAHAKLTKMEANFSE 113
L V+G++ PDE+ + + L++ +E + + EA + R + + +K + +FS+
Sbjct 1249 LATIVQGSQRKPDELERHVCAFLAEMEENIGSSMTTEAFLERLRWLSSSKFHRSATSFSD 1308
Query 114 DFKKSAEEIFAHSNCFTKRDL 134
F + +I + + CF + L
Sbjct 1309 YFGEVTSQIASRNFCFIREQL 1329
> cel:Y70C5C.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=985
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 36/80 (45%), Gaps = 1/80 (1%)
Query 52 STALLQCFVKGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANF 111
T LQ V+G K+ D V++ I+ L ++ + MP+ E A+L +
Sbjct 809 GTVALQILVQGPKS-VDHVLERIEAFLESVRKEIVEMPQEEFENRVSGLIAQLEEKPKTL 867
Query 112 SEDFKKSAEEIFAHSNCFTK 131
S FKK +EI FT+
Sbjct 868 SCRFKKFWDEIECRQYNFTR 887
> cel:C02G6.1 hypothetical protein; K01408 insulysin [EC:3.4.24.56]
Length=980
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 1/82 (1%)
Query 56 LQCFVKGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDF 115
L FV+G K+ D V++ I+ L ++ + MP+ E + A+L + S F
Sbjct 802 LNIFVQGPKS-VDYVLERIEVFLESVRKEIIEMPQDEFEKKVAGMIARLEEKPKTLSNRF 860
Query 116 KKSAEEIFAHSNCFTKRDLEVK 137
K+ +I F +R+ EVK
Sbjct 861 KRFWYQIECRQYDFARREKEVK 882
> ath:AT5G50920 CLPC1; CLPC1; ATP binding / ATP-dependent peptidase/
ATPase; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=929
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 26/53 (49%), Gaps = 0/53 (0%)
Query 67 PDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFKKSA 119
PD+ + +IDE S+ + A +PE KE K EA +DF+K+
Sbjct 482 PDKAIDLIDEAGSRVRLRHAQVPEEARELEKELRQITKEKNEAVRGQDFEKAG 534
> ath:AT1G12430 ARK3; ARK3 (ARMADILLO REPEAT KINESIN 3); ATP binding
/ binding / microtubule motor
Length=919
Score = 29.6 bits (65), Expect = 2.9, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 36/64 (56%), Gaps = 3/64 (4%)
Query 64 KAHPDEVVKMIDE---ELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFKKSAE 120
KA DE+ ++ E ++S+A++ AN E E R++ + K+E N+S++ KK A
Sbjct 447 KAFVDEIERITVEAHNQISEAEKRYANALEDEKLRYQNDYMESIKKLEENWSKNQKKLAA 506
Query 121 EIFA 124
E A
Sbjct 507 ERLA 510
> tgo:TGME49_053870 hypothetical protein
Length=4787
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 5 HAKLTKMEA--NFSEDFKKSAEEIFAHSNCFTKR 36
+ +L K+E FS DF KSA I H+ C KR
Sbjct 2244 YTRLGKLERLLRFSADFTKSAHHILLHAACLRKR 2277
Score = 29.3 bits (64), Expect = 3.5, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 101 HAKLTKMEA--NFSEDFKKSAEEIFAHSNCFTKR 132
+ +L K+E FS DF KSA I H+ C KR
Sbjct 2244 YTRLGKLERLLRFSADFTKSAHHILLHAACLRKR 2277
> dre:100332871 protein-kinase, interferon-inducible double stranded
RNA dependent inhibitor, repressor of (P58 repressor)-like
Length=2900
Score = 29.3 bits (64), Expect = 4.2, Method: Composition-based stats.
Identities = 17/44 (38%), Positives = 26/44 (59%), Gaps = 1/44 (2%)
Query 55 LLQCFVKGAKAHPDEVVKMIDEELSKAKEYLANMPEAEMARWKE 98
LLQ F + K E+ K +D L+ +E LA +PE ++RWK+
Sbjct 867 LLQFFDQSPKLG-SELAKTMDGLLNTPREALAEVPETCLSRWKK 909
> dre:768157 cplx4a, CPLX4, si:ch211-233a1.5, zgc:153429; complexin
4a
Length=159
Score = 28.9 bits (63), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 32/53 (60%), Gaps = 6/53 (11%)
Query 67 PDEVVKMIDEELSKAKE------YLANMPEAEMARWKEAAHAKLTKMEANFSE 113
P+E++KM+DE+ ++ +E + N+ +M + KE A A LT+M++ E
Sbjct 102 PEELLKMVDEDATEEEEKDSIMGQIQNLQNMDMDQIKEKASATLTEMKSKAEE 154
> ath:AT5G64070 PI-4KBETA1 (PHOSPHATIDYLINOSITOL 4-OH KINASE BETA1);
1-phosphatidylinositol 4-kinase; K00888 phosphatidylinositol
4-kinase [EC:2.7.1.67]
Length=1121
Score = 28.5 bits (62), Expect = 6.5, Method: Composition-based stats.
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 0/60 (0%)
Query 18 DFKKSAEEIFAHSNCFTKRDLEVKYLDNDFSQAASTALLQCFVKGAKAHPDEVVKMIDEE 77
D K EE+ A+S+ F KR L +++ S S + + +K D+V K +D+E
Sbjct 321 DNKGDEEELGANSDSFFKRLLRESKNEDEESNPNSEGFFKKLFRDSKPEDDKVPKEVDDE 380
> ath:AT3G48870 HSP93-III; ATP binding / ATPase/ DNA binding /
nuclease/ nucleoside-triphosphatase/ nucleotide binding / protein
binding; K03696 ATP-dependent Clp protease ATP-binding
subunit ClpC
Length=952
Score = 28.5 bits (62), Expect = 6.8, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 67 PDEVVKMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFK 116
PD+ + +IDE S+ + A +PE K+ K EA S+DF+
Sbjct 503 PDKAIDLIDEAGSRVRLRHAQLPEEARELEKQLRQITKEKNEAVRSQDFE 552
> ath:AT4G00955 hypothetical protein
Length=252
Score = 28.1 bits (61), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 27/70 (38%), Gaps = 3/70 (4%)
Query 7 KLTKMEANFSEDFKKSAEEIFAHSNCFTKRD---LEVKYLDNDFSQAASTALLQCFVKGA 63
K+ M F + F S+E F NC + + ++LD ST + CF
Sbjct 95 KIEDMTPLFGKQFTPSSENSFLMENCVNPTNGCSINQRFLDKQLKSCESTGNISCFPSDT 154
Query 64 KAHPDEVVKM 73
+ E + M
Sbjct 155 SSKSSEFLSM 164
> dre:559018 clul1, wu:fb51b10; clusterin-like 1 (retinal)
Length=466
Score = 27.7 bits (60), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Query 72 KMIDEELSKAKEYLANMPEAEMARWKEAAHAKLTKMEANFSEDFKKSAEEI 122
K++D+E+SKA + M E MAR E HA L K SE+ KK A EI
Sbjct 45 KLVDQEVSKALYGIKQMKEV-MAR-NEEKHANLMK-SLRHSEEKKKGAAEI 92
Lambda K H
0.314 0.124 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40