bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3127_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
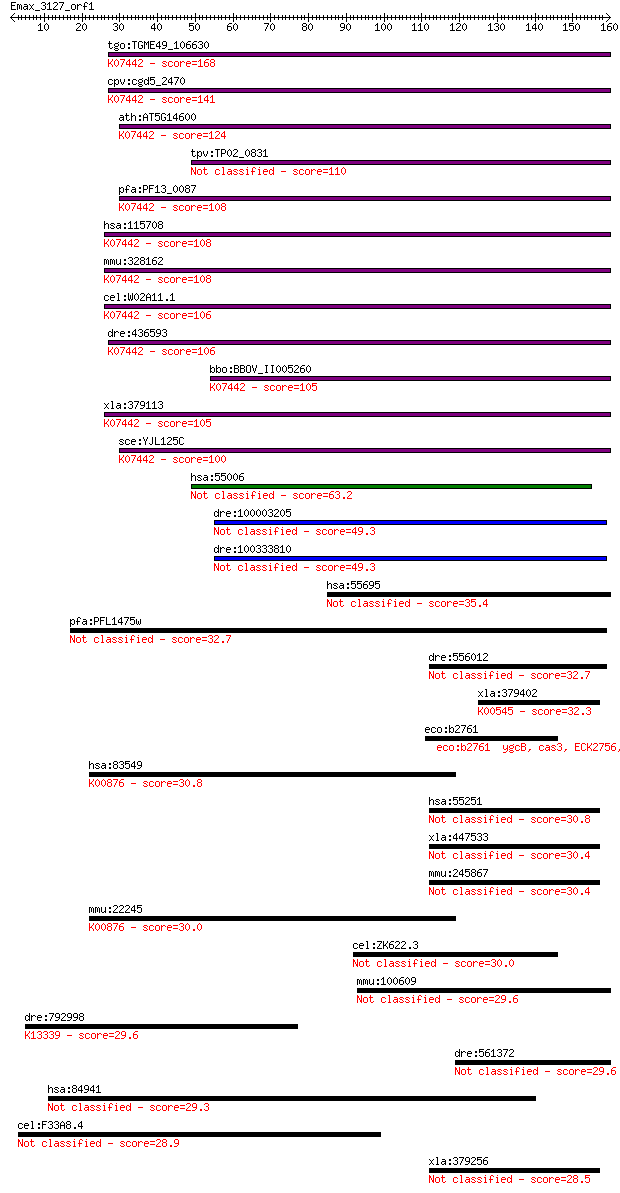
tgo:TGME49_106630 tRNA (adenine-N(1)-)-methyltransferase catal... 168 6e-42
cpv:cgd5_2470 GCD14 RNA methylase ; K07442 tRNA (adenine-N1-)-... 141 7e-34
ath:AT5G14600 tRNA (adenine-N1-)-methyltransferase; K07442 tRN... 124 1e-28
tpv:TP02_0831 hypothetical protein 110 2e-24
pfa:PF13_0087 methyltransferase, putative; K07442 tRNA (adenin... 108 5e-24
hsa:115708 TRMT61A, C14orf172, FLJ40452, GCD14, Gcd14p, TRM61,... 108 8e-24
mmu:328162 Trmt61a, 6720458F09Rik, AI606093, Gcd14, Trm61; tRN... 108 8e-24
cel:W02A11.1 hypothetical protein; K07442 tRNA (adenine-N1-)-m... 106 3e-23
dre:436593 trmt61a, zgc:86657; tRNA methyltransferase 61 homol... 106 3e-23
bbo:BBOV_II005260 18.m06436; hypothetical protein; K07442 tRNA... 105 4e-23
xla:379113 trmt61a, MGC53312; tRNA methyltransferase 61 homolo... 105 5e-23
sce:YJL125C GCD14, TRM61; 1-methyladenosine tRNA methyltransfe... 100 2e-21
hsa:55006 TRMT61B, DKFZp564I2178, FLJ20628; tRNA methyltransfe... 63.2 4e-10
dre:100003205 hypothetical LOC100003205 49.3 5e-06
dre:100333810 hypothetical protein LOC100333810 49.3 5e-06
hsa:55695 NSUN5, FLJ10267, MGC986, NOL1, NOL1R, NSUN5A, WBSCR2... 35.4 0.085
pfa:PFL1475w sun-family protein, putative 32.7 0.52
dre:556012 pcmtd2, MGC136935, zgc:136935; protein-L-isoasparta... 32.7 0.58
xla:379402 MGC53924; similar to catecholamine-O-methyltransfer... 32.3 0.60
eco:b2761 ygcB, cas3, ECK2756, JW2731; Cas3 predicted helicase... 31.2 1.6
hsa:83549 UCK1, FLJ12255, URK1; uridine-cytidine kinase 1 (EC:... 30.8 1.7
hsa:55251 PCMTD2, C20orf36, FLJ10883, KIAA0835, KIAA1050; prot... 30.8 2.0
xla:447533 MGC83799 protein 30.4 2.4
mmu:245867 Pcmtd2, 5330414D10Rik, AI314201, MGC36470; protein-... 30.4 2.6
mmu:22245 Uck1, URK1, Umpk; uridine-cytidine kinase 1 (EC:2.7.... 30.0 2.9
cel:ZK622.3 pmt-1; Phosphoethanolamine MethyTransferase family... 30.0 3.2
mmu:100609 Nsun5, 9830109N13Rik, AI326939, Nol1r, Wbscr20, Wbs... 29.6 3.8
dre:792998 si:ch73-197b13.2; K13339 peroxin-6 29.6 3.9
dre:561372 comta, MGC114157, zgc:114157; catechol-O-methyltran... 29.6 4.5
hsa:84941 HSH2D, ALX, FLJ14886, HSH2; hematopoietic SH2 domain... 29.3 5.3
cel:F33A8.4 hypothetical protein 28.9 7.7
xla:379256 pcmtd2, MGC53341; protein-L-isoaspartate (D-asparta... 28.5 9.0
> tgo:TGME49_106630 tRNA (adenine-N(1)-)-methyltransferase catalytic
subunit, putative (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=360
Score = 168 bits (426), Expect = 6e-42, Method: Compositional matrix adjust.
Identities = 77/133 (57%), Positives = 102/133 (76%), Gaps = 0/133 (0%)
Query 27 EFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLV 86
E+P+ G+ VILYGS +++I +L R + +RLGN+ H+DI+ T G KV +++ +WLV
Sbjct 76 EYPRYGDFVILYGSHQLVIPVLLERGRVSNSRLGNYRHDDIVTTAYGSKVQDRKSKRWLV 135
Query 87 VLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRP 146
VLRP+PDL +LAL HRTQI+YH+DISL+L LLD PG+ I EAGTGSGSLS LA A+ P
Sbjct 136 VLRPSPDLFSLALTHRTQILYHSDISLVLMLLDACPGRRICEAGTGSGSLSCHLARAVAP 195
Query 147 GGRLFTFEFHEQR 159
G +FTFEFH+QR
Sbjct 196 TGHIFTFEFHQQR 208
> cpv:cgd5_2470 GCD14 RNA methylase ; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=312
Score = 141 bits (356), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 64/133 (48%), Positives = 93/133 (69%), Gaps = 0/133 (0%)
Query 27 EFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLV 86
E K G+LV+L+G EM+I L + G +TR G F H +I+ G ++++ + KW+V
Sbjct 44 EMCKNGDLVVLFGGYEMVIQVFLEQNGRTQTRNGVFLHNEIIGKNFGSRIFDTSKSKWMV 103
Query 87 VLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRP 146
VL+ +P+L +++L HRTQI+Y ADISLI LLD PGK I+EAGTGSGSL+ SL + P
Sbjct 104 VLKLSPELISISLNHRTQILYQADISLICVLLDASPGKNIIEAGTGSGSLTVSLCRSTNP 163
Query 147 GGRLFTFEFHEQR 159
GG ++TFE+ ++R
Sbjct 164 GGAVYTFEYDQKR 176
> ath:AT5G14600 tRNA (adenine-N1-)-methyltransferase; K07442 tRNA
(adenine-N1-)-methyltransferase catalytic subunit [EC:2.1.1.36]
Length=318
Score = 124 bits (311), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 59/130 (45%), Positives = 91/130 (70%), Gaps = 1/130 (0%)
Query 30 KEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLVVLR 89
++G+LVI+Y +++ + + G+L+ R G + H D + +G KV+ + GK++ +L
Sbjct 17 EDGDLVIVYERHDVMKPVKVSKDGVLQNRFGVYKHSDWIGKPLGTKVFSNK-GKFVYLLA 75
Query 90 PTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGR 149
PTP+L TL L HRTQI+Y ADIS ++ L+V PG ++E+GTGSGSLS SLA A+ P G
Sbjct 76 PTPELWTLVLSHRTQILYIADISFVVMYLEVVPGCVVLESGTGSGSLSTSLARAVAPTGH 135
Query 150 LFTFEFHEQR 159
+++F+FHEQR
Sbjct 136 VYSFDFHEQR 145
> tpv:TP02_0831 hypothetical protein
Length=312
Score = 110 bits (274), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 52/113 (46%), Positives = 77/113 (68%), Gaps = 2/113 (1%)
Query 49 LRRAGILRTRLGNFSHEDIMKTRIGHKVY--EQRQGKWLVVLRPTPDLHTLALRHRTQII 106
++ ++ + G F + G K++ E + W+V+L+PTP+L T ++ HRTQI+
Sbjct 39 VKNKRLIHNKKGIFDLASCIGKFYGQKLFWDENNKKHWVVLLKPTPELITKSIIHRTQIL 98
Query 107 YHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFHEQR 159
Y ADISLI+ LLD+ PGK ++E GTGSGSLSY+LA ++ P G +FTF+FH QR
Sbjct 99 YRADISLIVLLLDLMPGKRVLECGTGSGSLSYALASSVAPNGHVFTFDFHPQR 151
> pfa:PF13_0087 methyltransferase, putative; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=316
Score = 108 bits (271), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 50/130 (38%), Positives = 78/130 (60%), Gaps = 0/130 (0%)
Query 30 KEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLVVLR 89
KEG V++Y E I L GI+ + G F H+DI+ G K+Y+ ++ VL+
Sbjct 3 KEGNCVVIYSDEENIDLLKLTNDGIINNKKGRFLHKDIIGKEYGSKIYDTLNKNYIYVLK 62
Query 90 PTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGR 149
+P+L L+ +TQ +Y DIS I + + P K I+EAGTG+G L+Y+LA ++ P G
Sbjct 63 RSPELIVNCLKKKTQTLYEHDISFICLVCNALPNKKILEAGTGTGCLTYALANSVLPNGV 122
Query 150 LFTFEFHEQR 159
+ TFE++E+R
Sbjct 123 IHTFEYNEER 132
> hsa:115708 TRMT61A, C14orf172, FLJ40452, GCD14, Gcd14p, TRM61,
hTRM61; tRNA methyltransferase 61 homolog A (S. cerevisiae)
(EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=289
Score = 108 bits (270), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 59/135 (43%), Positives = 83/135 (61%), Gaps = 2/135 (1%)
Query 26 EEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHE-DIMKTRIGHKVYEQRQGKW 84
EE KEG+ IL +++ ++R +TR G H D++ G KV R G W
Sbjct 7 EELIKEGDTAILSLGHGAMVAVRVQRGAQTQTRHGVLRHSVDLIGRPFGSKVTCGR-GGW 65
Query 85 LVVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMAL 144
+ VL PTP+L TL L HRTQI+Y DI+LI +L++RPG + E+GTGSGS+S+++ +
Sbjct 66 VYVLHPTPELWTLNLPHRTQILYSTDIALITMMLELRPGSVVCESGTGSGSVSHAIIRTI 125
Query 145 RPGGRLFTFEFHEQR 159
P G L T EFH+QR
Sbjct 126 APTGHLHTVEFHQQR 140
> mmu:328162 Trmt61a, 6720458F09Rik, AI606093, Gcd14, Trm61; tRNA
methyltransferase 61 homolog A (S. cerevisiae) (EC:2.1.1.36);
K07442 tRNA (adenine-N1-)-methyltransferase catalytic
subunit [EC:2.1.1.36]
Length=290
Score = 108 bits (270), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 58/135 (42%), Positives = 84/135 (62%), Gaps = 2/135 (1%)
Query 26 EEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHE-DIMKTRIGHKVYEQRQGKW 84
EE KEG+ IL +++ ++R +TR G H D++ G KV R G W
Sbjct 7 EELIKEGDTAILSLGHGSMVAVRVQRGAQTQTRHGVLRHSVDLIGRPFGSKVICSR-GGW 65
Query 85 LVVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMAL 144
+ VL PTP+L T+ L HRTQI+Y DI+LI +L++RPG + E+GTGSGS+S+++ ++
Sbjct 66 VYVLHPTPELWTVNLPHRTQILYSTDIALITMMLELRPGSVVCESGTGSGSVSHAIIRSV 125
Query 145 RPGGRLFTFEFHEQR 159
P G L T EFH+QR
Sbjct 126 APTGHLHTVEFHQQR 140
> cel:W02A11.1 hypothetical protein; K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=312
Score = 106 bits (265), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 55/134 (41%), Positives = 83/134 (61%), Gaps = 2/134 (1%)
Query 26 EEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWL 85
E+ +EG+ VI+Y + ++ +++R L + G HE I+ R G ++ ++
Sbjct 12 EDRIQEGDTVIVYVTYGQMVPTIVKRGQTLMMKYGALRHEFIIGKRWGQRL--SATAGYV 69
Query 86 VVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
+LRPT DL T AL RTQI+Y AD + ILSLLD +PG I E+GTGSGSLS+++A+ +
Sbjct 70 YILRPTSDLWTRALPRRTQILYTADCAQILSLLDAKPGSVICESGTGSGSLSHAIALTVA 129
Query 146 PGGRLFTFEFHEQR 159
P G L+T + E R
Sbjct 130 PTGHLYTHDIDETR 143
> dre:436593 trmt61a, zgc:86657; tRNA methyltransferase 61 homolog
A (S. cerevisiae) (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=305
Score = 106 bits (265), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 55/134 (41%), Positives = 83/134 (61%), Gaps = 2/134 (1%)
Query 27 EFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHE-DIMKTRIGHKVYEQRQGKWL 85
E +EG++VI++ + ++ ++ +TR G H D++ R G KV + G W+
Sbjct 8 ELIQEGDVVIIFMGHDSMMPIKVQSGAQTQTRYGVIRHSSDLIGQRFGSKVTCSK-GGWV 66
Query 86 VVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
VL PTP+L T+ L HRTQI+Y DI+ I +L+++PG + E+GTGSGSLS+S+ +
Sbjct 67 YVLHPTPELWTINLPHRTQILYTTDIANITLMLELKPGSVVCESGTGSGSLSHSILRTIA 126
Query 146 PGGRLFTFEFHEQR 159
P G L T EFH QR
Sbjct 127 PTGHLHTVEFHSQR 140
> bbo:BBOV_II005260 18.m06436; hypothetical protein; K07442 tRNA
(adenine-N1-)-methyltransferase catalytic subunit [EC:2.1.1.36]
Length=286
Score = 105 bits (263), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 53/107 (49%), Positives = 71/107 (66%), Gaps = 1/107 (0%)
Query 54 ILRTRLGNFSHEDIMKTRIGHKVY-EQRQGKWLVVLRPTPDLHTLALRHRTQIIYHADIS 112
IL + G F ++ + G K Y + +W V+L+PT +L T + HRTQI+Y ADIS
Sbjct 44 ILHRKGGIFDLRKVVGKQYGQKFYWDSVANRWSVILQPTGELITRTVTHRTQILYRADIS 103
Query 113 LILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFHEQR 159
L + LLD+ PGK ++E GTGSGSLSYSLA + P G L+TF+FH QR
Sbjct 104 LAILLLDLYPGKKVLECGTGSGSLSYSLATTVAPTGHLYTFDFHNQR 150
> xla:379113 trmt61a, MGC53312; tRNA methyltransferase 61 homolog
A; K07442 tRNA (adenine-N1-)-methyltransferase catalytic
subunit [EC:2.1.1.36]
Length=303
Score = 105 bits (263), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 51/135 (37%), Positives = 84/135 (62%), Gaps = 2/135 (1%)
Query 26 EEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHE-DIMKTRIGHKVYEQRQGKW 84
++ ++G+ I++ + + ++ +T+ G H D++ + G KV + G W
Sbjct 7 KDLIEDGDTAIVFLGHDSMFPITIQNGSQTQTKYGVIRHSTDLIGKKFGSKVTCSK-GGW 65
Query 85 LVVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMAL 144
+ +L PTP+L TL L HRTQI+Y DISLI +L+++PG + E+GTGSGSLS+++ +
Sbjct 66 VYILYPTPELWTLNLPHRTQILYSTDISLITMMLELKPGSVVCESGTGSGSLSHAIIRTV 125
Query 145 RPGGRLFTFEFHEQR 159
P G L+T EFH+QR
Sbjct 126 APTGHLYTVEFHQQR 140
> sce:YJL125C GCD14, TRM61; 1-methyladenosine tRNA methyltransferase
subunit (EC:2.1.1.36); K07442 tRNA (adenine-N1-)-methyltransferase
catalytic subunit [EC:2.1.1.36]
Length=383
Score = 100 bits (250), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 54/134 (40%), Positives = 83/134 (61%), Gaps = 7/134 (5%)
Query 30 KEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGK----WL 85
KEG+L +++ SR+ I + + TR G+F H+DI+ G ++ + +G ++
Sbjct 14 KEGDLTLIWVSRDNIKPVRMHSEEVFNTRYGSFPHKDIIGKPYGSQIAIRTKGSNKFAFV 73
Query 86 VVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
VL+PTP+L TL+L HRTQI+Y D S I+ L+ P ++EAGTGSGS S++ A ++
Sbjct 74 HVLQPTPELWTLSLPHRTQIVYTPDSSYIMQRLNCSPHSRVIEAGTGSGSFSHAFARSV- 132
Query 146 PGGRLFTFEFHEQR 159
G LF+FEFH R
Sbjct 133 --GHLFSFEFHHIR 144
> hsa:55006 TRMT61B, DKFZp564I2178, FLJ20628; tRNA methyltransferase
61 homolog B (S. cerevisiae) (EC:2.1.1.36)
Length=477
Score = 63.2 bits (152), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 60/106 (56%), Gaps = 1/106 (0%)
Query 49 LRRAGILRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLVVLRPTPDLHTLALRHRTQIIYH 108
L G+L + G I+ G ++ GK ++ RP + + + ++ T I +
Sbjct 174 LNNFGLLNSNWGAVPFGKIVGKFPG-QILRSSFGKQYMLRRPALEDYVVLMKRGTAITFP 232
Query 109 ADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFE 154
DI++ILS++D+ PG T++EAG+GSG +S L+ A+ GR+ +FE
Sbjct 233 KDINMILSMMDINPGDTVLEAGSGSGGMSLFLSKAVGSQGRVISFE 278
> dre:100003205 hypothetical LOC100003205
Length=450
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 53/104 (50%), Gaps = 1/104 (0%)
Query 55 LRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLVVLRPTPDLHTLALRHRTQIIYHADISLI 114
L + G+ +H+D M+ R V +G V+ R + + L ++ I Y D S +
Sbjct 166 LYSNWGSVAHDD-MEGRPAGSVIRTSKGIPFVIRRVSLEEFVLFMKRGPAIAYPKDASAM 224
Query 115 LSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFHEQ 158
L+++DV G ++E G+GSG +S L+ A+ G + + E E
Sbjct 225 LTMMDVTEGDCVLECGSGSGGMSLFLSRAVGYKGSVLSVEIRED 268
> dre:100333810 hypothetical protein LOC100333810
Length=445
Score = 49.3 bits (116), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 30/104 (28%), Positives = 53/104 (50%), Gaps = 1/104 (0%)
Query 55 LRTRLGNFSHEDIMKTRIGHKVYEQRQGKWLVVLRPTPDLHTLALRHRTQIIYHADISLI 114
L + G+ +H+D M+ R V +G V+ R + + L ++ I Y D S +
Sbjct 161 LYSNWGSVAHDD-MEGRPAGSVIRTSKGIPFVIRRVSLEEFVLFMKRGPAIAYPKDASAM 219
Query 115 LSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFHEQ 158
L+++DV G ++E G+GSG +S L+ A+ G + + E E
Sbjct 220 LTMMDVTKGDCVLECGSGSGGMSLFLSRAVGYKGSVLSVEIRED 263
> hsa:55695 NSUN5, FLJ10267, MGC986, NOL1, NOL1R, NSUN5A, WBSCR20,
WBSCR20A, p120; NOP2/Sun domain family, member 5
Length=470
Score = 35.4 bits (80), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Query 85 LVVLRPTPDLHTLALRHRTQIIYHADISLILS-LLDVRPGKTIVEAGTGSGSLSYSLAMA 143
L+V DLH L +I S + + LLD PG +++A G+ + LA
Sbjct 188 LLVFPAQTDLHEHPLYRAGHLILQDRASCLPAMLLDPPPGSHVIDACAAPGNKTSHLAAL 247
Query 144 LRPGGRLFTFEFHEQR 159
L+ G++F F+ +R
Sbjct 248 LKNQGKIFAFDLDAKR 263
> pfa:PFL1475w sun-family protein, putative
Length=376
Score = 32.7 bits (73), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 35/150 (23%), Positives = 70/150 (46%), Gaps = 10/150 (6%)
Query 17 QLSAHLQQSEEFPKE--GELVILYGSREMI--ISCVLRRAGI-LRTRLGNFSHEDIMKTR 71
++ AH++ S FPKE +L+ YG ++ I +S + +A + LR S ++ KT
Sbjct 88 KIPAHVRYS--FPKELYNQLLDCYGEKKSITLMSILNEKAPVFLRVNNNKISRNELYKTL 145
Query 72 I--GHKVYEQRQGKWLVVLRPTPDLHTLALRHRTQIIYHADISLILS-LLDVRPGKTIVE 128
I G V + + ++L L + + + S I+S + V PG +++
Sbjct 146 ISKGISVEKCVNSPYGLLLTKNQILKNIHEYKKGYFEIQDEASQIVSSKIPVHPGDKVLD 205
Query 129 AGTGSGSLSYSLAMALRPGGRLFTFEFHEQ 158
GSG + + +M + G+++ + +Q
Sbjct 206 YCAGSGGKTLAFSMLMENTGKIYLHDIRDQ 235
> dre:556012 pcmtd2, MGC136935, zgc:136935; protein-L-isoaspartate
(D-aspartate) O-methyltransferase domain containing 2
Length=376
Score = 32.7 bits (73), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 112 SLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFHEQ 158
S ++ LD++PG + + G+G+G LS + + L P G E HE
Sbjct 70 SEVMEALDLQPGLSFLNLGSGTGYLSTMVGLILGPFGVNHGVELHED 116
> xla:379402 MGC53924; similar to catecholamine-O-methyltransferase;
K00545 catechol O-methyltransferase [EC:2.1.1.6]
Length=261
Score = 32.3 bits (72), Expect = 0.60, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 125 TIVEAGTGSGSLSYSLAMALRPGGRLFTFEFH 156
T++E GT G + + +L+PG R FT EF+
Sbjct 108 TLLELGTYCGYSAIRICRSLKPGARFFTVEFN 139
> eco:b2761 ygcB, cas3, ECK2756, JW2731; Cas3 predicted helicase
needed for Cascade anti-viral activity; K07012
Length=888
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 111 ISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
+ +++ L V PG T++EA TGSG +LA A +
Sbjct 296 LQVLVDALPVAPGLTVIEAPTGSGKTETALAYAWK 330
> hsa:83549 UCK1, FLJ12255, URK1; uridine-cytidine kinase 1 (EC:2.7.1.48);
K00876 uridine kinase [EC:2.7.1.48]
Length=201
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 27/98 (27%), Positives = 47/98 (47%), Gaps = 4/98 (4%)
Query 22 LQQSEEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQ-R 80
L Q+E ++ ++VIL R + ++A L+ + NF H D + H+ +
Sbjct 47 LGQNEVEQRQRKVVILSQDRFYKVLTAEQKAKALKGQY-NFDHPDAFDNDLMHRTLKNIV 105
Query 81 QGKWLVVLRPTPDLHTLALRHRTQIIYHADISLILSLL 118
+GK + V PT D T + T ++Y AD+ L +L
Sbjct 106 EGKTVEV--PTYDFVTHSRLPETTVVYPADVVLFEGIL 141
> hsa:55251 PCMTD2, C20orf36, FLJ10883, KIAA0835, KIAA1050; protein-L-isoaspartate
(D-aspartate) O-methyltransferase domain
containing 2
Length=334
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 112 SLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFH 156
S ++ LD++PG + + G+G+G LS + + L P G E H
Sbjct 70 SEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILGPFGVNHGVELH 114
> xla:447533 MGC83799 protein
Length=361
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 112 SLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFH 156
S ++ LD++PG + + G+G+G LS + + L P G E H
Sbjct 70 SEVMEALDLQPGLSFLNLGSGTGYLSTMVGLILGPFGINHGVEIH 114
> mmu:245867 Pcmtd2, 5330414D10Rik, AI314201, MGC36470; protein-L-isoaspartate
(D-aspartate) O-methyltransferase domain containing
2
Length=359
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 112 SLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFH 156
S ++ LD++PG + + G+G+G LS + + L P G E H
Sbjct 70 SEVMEALDLQPGLSFLNLGSGTGYLSSMVGLILGPFGVNHGVELH 114
> mmu:22245 Uck1, URK1, Umpk; uridine-cytidine kinase 1 (EC:2.7.1.48);
K00876 uridine kinase [EC:2.7.1.48]
Length=283
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 46/98 (46%), Gaps = 4/98 (4%)
Query 22 LQQSEEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKTRIGHKVYEQ-R 80
L Q+E ++ +LVIL + ++A L+ + NF H D + HK +
Sbjct 53 LGQNEVDRRQRKLVILSQDCFYKVLTAEQKAKALKGQY-NFDHPDAFDNDLMHKTLKNIV 111
Query 81 QGKWLVVLRPTPDLHTLALRHRTQIIYHADISLILSLL 118
+GK + V PT D T + T ++Y AD+ L +L
Sbjct 112 EGKTVEV--PTYDFVTHSRLPETTVVYPADVVLFEGIL 147
> cel:ZK622.3 pmt-1; Phosphoethanolamine MethyTransferase family
member (pmt-1)
Length=475
Score = 30.0 bits (66), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 92 PDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALR 145
PD +++ L H + + +D + IL+ L + K +V+ G G G + LA R
Sbjct 35 PDTNSMMLNHSAEELESSDRADILASLPLLHNKDVVDIGAGIGRFTTVLAETAR 88
> mmu:100609 Nsun5, 9830109N13Rik, AI326939, Nol1r, Wbscr20, Wbscr20a;
NOL1/NOP2/Sun domain family, member 5
Length=465
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 32/68 (47%), Gaps = 1/68 (1%)
Query 93 DLHTLALRHRTQIIYHADISLILS-LLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLF 151
DLH L +I S + + LL PG +++A G+ + +A L+ G++F
Sbjct 196 DLHEHPLYRAGHLILQDKASCLPAMLLSPPPGSHVIDACAAPGNKTSYIAALLKNQGKIF 255
Query 152 TFEFHEQR 159
F+ +R
Sbjct 256 AFDQDAKR 263
> dre:792998 si:ch73-197b13.2; K13339 peroxin-6
Length=956
Score = 29.6 bits (65), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 35/72 (48%), Gaps = 2/72 (2%)
Query 5 FGVFVEAEESAFQLSAHLQQSEEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSH 64
F F+EA E +L + ++ + K G +V E+ + C + + L +LG F+
Sbjct 242 FSAFIEALEC--RLDVRVVETSDLLKPGGIVSALNEEEVDVDCCVFVSRSLLVKLGVFNG 299
Query 65 EDIMKTRIGHKV 76
E ++ + G KV
Sbjct 300 EWVIASVPGEKV 311
> dre:561372 comta, MGC114157, zgc:114157; catechol-O-methyltransferase
a
Length=143
Score = 29.6 bits (65), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 1/41 (2%)
Query 119 DVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFHEQR 159
+V P T +E GT G + +A L PG +L T EF+ R
Sbjct 98 EVNPS-TALELGTYCGYSTVRIARLLSPGTKLITLEFNPDR 137
> hsa:84941 HSH2D, ALX, FLJ14886, HSH2; hematopoietic SH2 domain
containing
Length=352
Score = 29.3 bits (64), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 54/129 (41%), Gaps = 19/129 (14%)
Query 11 AEESAFQLSAHLQQSEEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNFSHEDIMKT 70
AEE+A +SA E PK V+ + S+E S + R + T+ S K+
Sbjct 146 AEEAACPVSA---PEEASPKP---VLCHQSKERKPSAEMNR---ITTKEATSSCPP--KS 194
Query 71 RIGHKVYEQRQGKWLVVLRPTPDLHTLALRHRTQIIYHADISLILSLLDVRPGKTIVEAG 130
+G E RQ W + L R R Q+ H + SLLDVR I G
Sbjct 195 PLG----ETRQKLWRSLKM----LPERGQRVRQQLKSHLATVNLSSLLDVRRSTVISGPG 246
Query 131 TGSGSLSYS 139
TG GS +S
Sbjct 247 TGKGSQDHS 255
> cel:F33A8.4 hypothetical protein
Length=467
Score = 28.9 bits (63), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 3 PPFGVFVEAEESAFQLSAHLQQSEEFPKEGELVILYGSREMIISCVLRRAGILRTRLGNF 62
PPFGVF+E L + + + F G+ L+ S MI+ + R +L GNF
Sbjct 310 PPFGVFMEP-----LLKSIEKMKKRFVSTGKEETLFYS--MIVLPIYIRKYVLH---GNF 359
Query 63 SHEDIMKTRIGHKVYEQRQGKWLVVLRPTP----DLHTLA 98
D T GHK+Y+ + + + P DL +A
Sbjct 360 WMSDYRVTYEGHKLYQHSEKTIVRLFTDLPVECIDLKNVA 399
> xla:379256 pcmtd2, MGC53341; protein-L-isoaspartate (D-aspartate)
O-methyltransferase domain containing 2
Length=367
Score = 28.5 bits (62), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 112 SLILSLLDVRPGKTIVEAGTGSGSLSYSLAMALRPGGRLFTFEFH 156
S ++ LD++PG + + G+G+G L+ + L P G E H
Sbjct 70 SEVMEALDLQPGLSFLNLGSGTGYLNTMAGLILGPFGINHGVEIH 114
Lambda K H
0.323 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3647184800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40