bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3174_orf1
Length=107
Score E
Sequences producing significant alignments: (Bits) Value
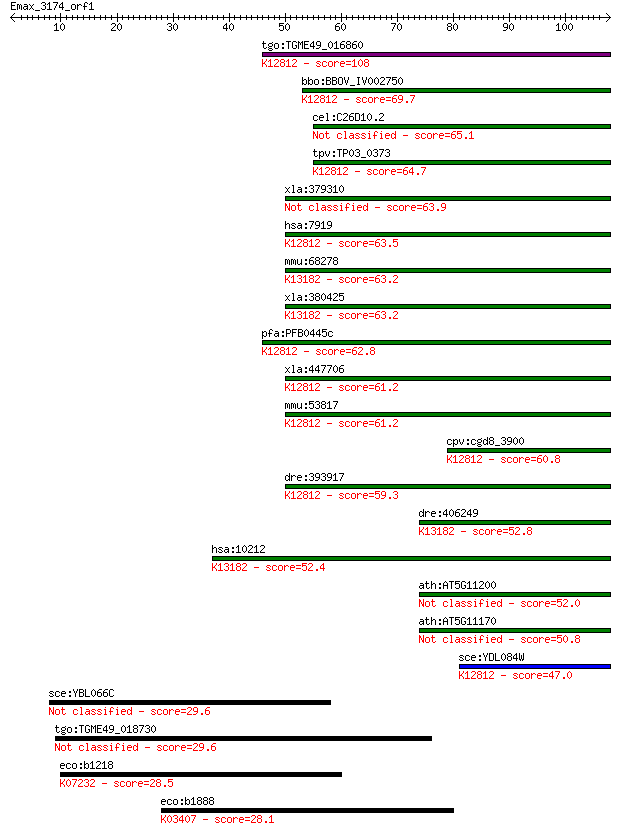
tgo:TGME49_016860 ATP-dependent RNA helicase, putative ; K1281... 108 6e-24
bbo:BBOV_IV002750 21.m02887; eIF-4A-like DEAD family RNA helic... 69.7 2e-12
cel:C26D10.2 hel-1; HELicase family member (hel-1) 65.1 5e-11
tpv:TP03_0373 ATP-dependent RNA helicase; K12812 ATP-dependent... 64.7 6e-11
xla:379310 ddx39a, MGC130793, MGC53693, bat1, bat1l, ddx39, dd... 63.9 1e-10
hsa:7919 DDX39B, BAT1, D6S81E, UAP56; DEAD (Asp-Glu-Ala-Asp) b... 63.5 1e-10
mmu:68278 Ddx39, 2610307C23Rik, BAT1, DDXL, Ddx39a, URH49; DEA... 63.2 2e-10
xla:380425 ddx39, MGC53944; nuclear RNA helicase; K13182 ATP-d... 63.2 2e-10
pfa:PFB0445c UAP56, U52; DEAD box helicase, UAP56; K12812 ATP-... 62.8 2e-10
xla:447706 ddx39b, MGC81606, bat1, uap56; DEAD (Asp-Glu-Ala-As... 61.2 7e-10
mmu:53817 Ddx39b, 0610030D10Rik, AI428441, Bat-1, Bat1, Bat1a,... 61.2 7e-10
cpv:cgd8_3900 Sub2p like superfamily II helicase involved in s... 60.8 9e-10
dre:393917 bat1, Bat1a, MGC63773, zgc:63773; HLA-B associated ... 59.3 3e-09
dre:406249 ddx39b, ddx39, wu:fc16a02, zgc:55433, zgc:85646; DE... 52.8 3e-07
hsa:10212 DDX39A, BAT1, BAT1L, DDX39, DDXL, MGC18203, MGC8417,... 52.4 3e-07
ath:AT5G11200 DEAD/DEAH box helicase, putative 52.0 5e-07
ath:AT5G11170 ATP binding / ATP-dependent helicase/ helicase/ ... 50.8 1e-06
sce:YDL084W SUB2; Component of the TREX complex required for n... 47.0 1e-05
sce:YBL066C SEF1; Putative transcription factor, has homolog i... 29.6 2.6
tgo:TGME49_018730 hypothetical protein 29.6 2.7
eco:b1218 chaC, ECK1212, JW1209; cation transport regulator; K... 28.5 4.9
eco:b1888 cheA, ECK1889, JW1877; fused chemotactic sensory his... 28.1 7.5
> tgo:TGME49_016860 ATP-dependent RNA helicase, putative ; K12812
ATP-dependent RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=434
Score = 108 bits (269), Expect = 6e-24, Method: Composition-based stats.
Identities = 53/62 (85%), Positives = 54/62 (87%), Gaps = 1/62 (1%)
Query 46 MAALEQNPADELVDYEEDEQQNDTKEKGVGEEAVGRGNYVSIHASGFRDFLLKPELLRAI 105
M LEQNPADELVDYEEDEQ ND KEKGV + VGRGNYVSIHASGFRDF LKPELLRAI
Sbjct 1 MTTLEQNPADELVDYEEDEQ-NDAKEKGVEDVVVGRGNYVSIHASGFRDFFLKPELLRAI 59
Query 106 GD 107
GD
Sbjct 60 GD 61
> bbo:BBOV_IV002750 21.m02887; eIF-4A-like DEAD family RNA helicase
(EC:3.6.1.3); K12812 ATP-dependent RNA helicase UAP56/SUB2
[EC:3.6.4.13]
Length=472
Score = 69.7 bits (169), Expect = 2e-12, Method: Composition-based stats.
Identities = 37/65 (56%), Positives = 43/65 (66%), Gaps = 10/65 (15%)
Query 53 PADELVDYEEDEQQNDTKEK--------GVGEE--AVGRGNYVSIHASGFRDFLLKPELL 102
PA++LVDYEE E T VG E +GRG+YV+IHASGFRDF LKPE+L
Sbjct 9 PAEDLVDYEEQEAGQVTSHNVIKSGASSKVGREDGTMGRGSYVAIHASGFRDFFLKPEIL 68
Query 103 RAIGD 107
RAIGD
Sbjct 69 RAIGD 73
> cel:C26D10.2 hel-1; HELicase family member (hel-1)
Length=425
Score = 65.1 bits (157), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 42/55 (76%), Gaps = 2/55 (3%)
Query 55 DELVDYEEDEQ--QNDTKEKGVGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
++L+DYEE+++ Q+ E G G+ +G Y SIH+SGFRDFLLKPE+LRAIGD
Sbjct 4 EQLLDYEEEQEEIQDKQPEVGGGDARKTKGTYASIHSSGFRDFLLKPEILRAIGD 58
> tpv:TP03_0373 ATP-dependent RNA helicase; K12812 ATP-dependent
RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=451
Score = 64.7 bits (156), Expect = 6e-11, Method: Composition-based stats.
Identities = 33/63 (52%), Positives = 42/63 (66%), Gaps = 10/63 (15%)
Query 55 DELVDYEEDEQQNDTKEKGVGEEA----------VGRGNYVSIHASGFRDFLLKPELLRA 104
++LVDYEE+E + K V + A +GRG+YV+IHASGFRDF LKPE+LRA
Sbjct 9 EDLVDYEEEEGETKVSTKDVIKSAAGKKSKTDGTMGRGSYVAIHASGFRDFFLKPEILRA 68
Query 105 IGD 107
I D
Sbjct 69 ISD 71
> xla:379310 ddx39a, MGC130793, MGC53693, bat1, bat1l, ddx39,
ddxl, dxd39; DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A
Length=427
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 34/60 (56%), Positives = 44/60 (73%), Gaps = 3/60 (5%)
Query 50 EQNPADELVDYEEDEQQNDTKEKGV--GEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
EQ+ +EL+DYEED++ E V + V +G+YVSIH+SGFRDFLLKPELLR+I D
Sbjct 3 EQDVENELLDYEEDDEPQAPAETAVPIARKEV-KGSYVSIHSSGFRDFLLKPELLRSIVD 61
> hsa:7919 DDX39B, BAT1, D6S81E, UAP56; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 39B (EC:3.6.4.13); K12812 ATP-dependent RNA
helicase UAP56/SUB2 [EC:3.6.4.13]
Length=428
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 36/62 (58%), Positives = 44/62 (70%), Gaps = 6/62 (9%)
Query 50 EQNPADELVDYEEDEQQNDTKEKGVGEEAVGR----GNYVSIHASGFRDFLLKPELLRAI 105
E + +EL+DYE+DE + T G G EA + G+YVSIH+SGFRDFLLKPELLRAI
Sbjct 3 ENDVDNELLDYEDDEVE--TAAGGDGAEAPAKKDVKGSYVSIHSSGFRDFLLKPELLRAI 60
Query 106 GD 107
D
Sbjct 61 VD 62
> mmu:68278 Ddx39, 2610307C23Rik, BAT1, DDXL, Ddx39a, URH49; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 39 (EC:3.6.4.13); K13182
ATP-dependent RNA helicase DDX39 [EC:3.6.4.13]
Length=427
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 33/59 (55%), Positives = 42/59 (71%), Gaps = 1/59 (1%)
Query 50 EQNPADELVDYEEDEQQNDTKEKGVGEEAVG-RGNYVSIHASGFRDFLLKPELLRAIGD 107
EQ+ +EL+DY+EDE+ +E +G+YVSIH+SGFRDFLLKPELLRAI D
Sbjct 3 EQDVENELLDYDEDEEPQAPQESTPAPPKKDVKGSYVSIHSSGFRDFLLKPELLRAIVD 61
> xla:380425 ddx39, MGC53944; nuclear RNA helicase; K13182 ATP-dependent
RNA helicase DDX39 [EC:3.6.4.13]
Length=427
Score = 63.2 bits (152), Expect = 2e-10, Method: Composition-based stats.
Identities = 33/60 (55%), Positives = 44/60 (73%), Gaps = 3/60 (5%)
Query 50 EQNPADELVDYEEDEQQNDTKEKG--VGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
EQ+ +EL+DYEED++ E + + V +G+YVSIH+SGFRDFLLKPELLR+I D
Sbjct 3 EQDVENELLDYEEDDEPQAPAETAAPIARKEV-KGSYVSIHSSGFRDFLLKPELLRSIVD 61
> pfa:PFB0445c UAP56, U52; DEAD box helicase, UAP56; K12812 ATP-dependent
RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=457
Score = 62.8 bits (151), Expect = 2e-10, Method: Composition-based stats.
Identities = 31/72 (43%), Positives = 43/72 (59%), Gaps = 10/72 (13%)
Query 46 MAALEQNPADELVDYEEDEQQNDTKE----------KGVGEEAVGRGNYVSIHASGFRDF 95
MA+++ N DELVDYE+DE D+K+ + RG+Y ++H GF+DF
Sbjct 1 MASMDHNAQDELVDYEDDENILDSKDVKGNLGNNILNNNNKGGAMRGSYATVHTGGFKDF 60
Query 96 LLKPELLRAIGD 107
LKPELLRAI +
Sbjct 61 FLKPELLRAISE 72
> xla:447706 ddx39b, MGC81606, bat1, uap56; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 39B; K12812 ATP-dependent RNA helicase
UAP56/SUB2 [EC:3.6.4.13]
Length=428
Score = 61.2 bits (147), Expect = 7e-10, Method: Composition-based stats.
Identities = 34/62 (54%), Positives = 44/62 (70%), Gaps = 6/62 (9%)
Query 50 EQNPADELVDYEEDEQQN----DTKEKGVGEEAVGRGNYVSIHASGFRDFLLKPELLRAI 105
E + +EL+DYE+D+ N D + V +E +G+YVSIH+SGFRDFLLKPELLRAI
Sbjct 3 ETDVDNELLDYEDDDVDNQAGVDAPDITVKKEM--KGSYVSIHSSGFRDFLLKPELLRAI 60
Query 106 GD 107
D
Sbjct 61 VD 62
> mmu:53817 Ddx39b, 0610030D10Rik, AI428441, Bat-1, Bat1, Bat1a,
D17H6S81E, D17H6S81E-1, D6S81Eh, MGC19235, MGC38799; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 39B (EC:3.6.4.13); K12812
ATP-dependent RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=428
Score = 61.2 bits (147), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 33/60 (55%), Positives = 41/60 (68%), Gaps = 2/60 (3%)
Query 50 EQNPADELVDYEEDEQQNDTKEKGVGEEAVG--RGNYVSIHASGFRDFLLKPELLRAIGD 107
E + +EL+DYE+DE + G A +G+YVSIH+SGFRDFLLKPELLRAI D
Sbjct 3 ENDVDNELLDYEDDEVETAAGADGTEAPAKKDVKGSYVSIHSSGFRDFLLKPELLRAIVD 62
> cpv:cgd8_3900 Sub2p like superfamily II helicase involved in
snRNP biogenesis ; K12812 ATP-dependent RNA helicase UAP56/SUB2
[EC:3.6.4.13]
Length=430
Score = 60.8 bits (146), Expect = 9e-10, Method: Composition-based stats.
Identities = 26/29 (89%), Positives = 28/29 (96%), Gaps = 0/29 (0%)
Query 79 VGRGNYVSIHASGFRDFLLKPELLRAIGD 107
VGRGNYV+IHASGFRDF LKPEL+RAIGD
Sbjct 36 VGRGNYVAIHASGFRDFFLKPELIRAIGD 64
> dre:393917 bat1, Bat1a, MGC63773, zgc:63773; HLA-B associated
transcript 1 (EC:3.6.1.-); K12812 ATP-dependent RNA helicase
UAP56/SUB2 [EC:3.6.4.13]
Length=435
Score = 59.3 bits (142), Expect = 3e-09, Method: Composition-based stats.
Identities = 33/67 (49%), Positives = 40/67 (59%), Gaps = 9/67 (13%)
Query 50 EQNPADELVDYEEDEQQNDTKEKGVGEEAVG---------RGNYVSIHASGFRDFLLKPE 100
E + +EL+DYEEDE + G +G+YVSIH+SGFRDFLLKPE
Sbjct 3 ENDVDNELLDYEEDEVDAGGAGDAGLGHSDGIISIRKEGVKGSYVSIHSSGFRDFLLKPE 62
Query 101 LLRAIGD 107
LLRAI D
Sbjct 63 LLRAIVD 69
> dre:406249 ddx39b, ddx39, wu:fc16a02, zgc:55433, zgc:85646;
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39b (EC:3.6.4.13); K13182
ATP-dependent RNA helicase DDX39 [EC:3.6.4.13]
Length=427
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 26/34 (76%), Positives = 31/34 (91%), Gaps = 1/34 (2%)
Query 74 VGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
VG++ V +G+YVSIH+SGFRDFLLKPELLRAI D
Sbjct 29 VGKKEV-KGSYVSIHSSGFRDFLLKPELLRAIVD 61
> hsa:10212 DDX39A, BAT1, BAT1L, DDX39, DDXL, MGC18203, MGC8417,
URH49; DEAD (Asp-Glu-Ala-Asp) box polypeptide 39A (EC:3.6.4.13);
K13182 ATP-dependent RNA helicase DDX39 [EC:3.6.4.13]
Length=470
Score = 52.4 bits (124), Expect = 3e-07, Method: Composition-based stats.
Identities = 35/72 (48%), Positives = 47/72 (65%), Gaps = 3/72 (4%)
Query 37 PPQTPPLFKMAALEQNPADELVDYEEDEQQNDTKEKGVGEEAVG-RGNYVSIHASGFRDF 95
PP + MA EQ+ ++L+DY+E+E+ +E +G+YVSIH+SGFRDF
Sbjct 35 PPSSWDYSIMA--EQDVENDLLDYDEEEEPQAPQESTPAPPKKDIKGSYVSIHSSGFRDF 92
Query 96 LLKPELLRAIGD 107
LLKPELLRAI D
Sbjct 93 LLKPELLRAIVD 104
> ath:AT5G11200 DEAD/DEAH box helicase, putative
Length=486
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 26/34 (76%), Positives = 28/34 (82%), Gaps = 1/34 (2%)
Query 74 VGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
V EAV +G YV IH+SGFRDFLLKPELLRAI D
Sbjct 31 VNGEAVKKG-YVGIHSSGFRDFLLKPELLRAIVD 63
> ath:AT5G11170 ATP binding / ATP-dependent helicase/ helicase/
nucleic acid binding
Length=427
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 26/34 (76%), Positives = 28/34 (82%), Gaps = 1/34 (2%)
Query 74 VGEEAVGRGNYVSIHASGFRDFLLKPELLRAIGD 107
V EAV +G YV IH+SGFRDFLLKPELLRAI D
Sbjct 31 VNGEAVKKG-YVGIHSSGFRDFLLKPELLRAIVD 63
> sce:YDL084W SUB2; Component of the TREX complex required for
nuclear mRNA export; member of the DEAD-box RNA helicase superfamily
and is involved in early and late steps of spliceosome
assembly; homolog of the human splicing factor hUAP56 (EC:3.6.1.-);
K12812 ATP-dependent RNA helicase UAP56/SUB2 [EC:3.6.4.13]
Length=446
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 19/27 (70%), Positives = 24/27 (88%), Gaps = 0/27 (0%)
Query 81 RGNYVSIHASGFRDFLLKPELLRAIGD 107
+G+YV IH++GF+DFLLKPEL RAI D
Sbjct 53 KGSYVGIHSTGFKDFLLKPELSRAIID 79
> sce:YBL066C SEF1; Putative transcription factor, has homolog
in Kluyveromyces lactis
Length=1148
Score = 29.6 bits (65), Expect = 2.6, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query 8 NRCETTFALLASSLAQEAAATAATAAVAPPPQTPPLFKMAALEQNPADEL 57
N + L ASSLAQ++ AT P + PPL +AL N + L
Sbjct 205 NEASSHMTLRASSLAQDSKGLVATE----PNKLPPLLNDSALPNNSKESL 250
> tgo:TGME49_018730 hypothetical protein
Length=256
Score = 29.6 bits (65), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 37/70 (52%), Gaps = 4/70 (5%)
Query 9 RCETTFALLASSLAQEAAATAATAAVAPPPQTPPLFKMAALEQ---NPADELVDYEEDEQ 65
R ET+ LL S+ + T++T AV P++ ++AA Q N DE + D +
Sbjct 161 RTETSRELL-QSVRSHSLTTSSTVAVTMSPRSSGTQQVAAGGQTTRNGGDERQEKHGDSR 219
Query 66 QNDTKEKGVG 75
Q++T+ GVG
Sbjct 220 QDETQTTGVG 229
> eco:b1218 chaC, ECK1212, JW1209; cation transport regulator;
K07232 cation transport protein ChaC
Length=231
Score = 28.5 bits (62), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 25/54 (46%), Gaps = 4/54 (7%)
Query 10 CETTFALLASSLA----QEAAATAATAAVAPPPQTPPLFKMAALEQNPADELVD 59
C+T F + SL Q AA+ AAT A P +F +L NPA E +
Sbjct 12 CKTAFGAIEESLLWSAEQRAASLAATLACRPDEGPVWIFGYGSLMWNPALEFTE 65
> eco:b1888 cheA, ECK1889, JW1877; fused chemotactic sensory histidine
kinase in two-component regulatory system with CheB
and CheY: sensory histidine kinase/signal sensing protein;
K03407 two-component system, chemotaxis family, sensor kinase
CheA [EC:2.7.13.3]
Length=654
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 28 TAATAAVAPPPQTPPLFKMAALEQNPADELVDYEEDEQQNDTKEKGVGEEAV 79
T T V+P TPP+ K+AA EQ P V+ E+ + N++ V E V
Sbjct 222 TFETVEVSPKISTPPVLKLAA-EQAPTGR-VEREKTTRSNESTSIRVAVEKV 271
Lambda K H
0.311 0.128 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007980300
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40