bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3194_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
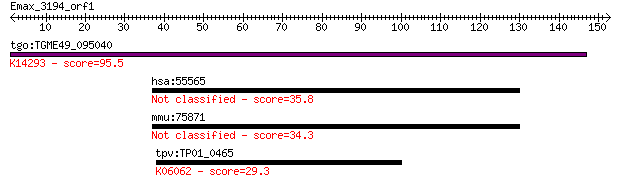
tgo:TGME49_095040 importin subunit beta-1, putative ; K14293 i... 95.5 5e-20
hsa:55565 ZNF821, FLJ16350; zinc finger protein 821 35.8
mmu:75871 Zfp821, 4930566A11Rik, Znf821; zinc finger protein 821 34.3
tpv:TP01_0465 histone acetyltransferase Gcn5; K06062 histone a... 29.3 5.3
> tgo:TGME49_095040 importin subunit beta-1, putative ; K14293
importin subunit beta-1
Length=971
Score = 95.5 bits (236), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 67/148 (45%), Positives = 90/148 (60%), Gaps = 12/148 (8%)
Query 1 VIQILCLRLGDQVKPVADTLWSTLKKVFQR--PANGVQPSQAEGSTSPNNPQGFVVSSEP 58
VIQILCLRLGDQV+PVA +W+ L ++F+ PAN P G S + +SE
Sbjct 636 VIQILCLRLGDQVQPVASQIWACLARIFRGSPPANA--PPGTAGQLSCS-------ASES 686
Query 59 SVCDALLALSALINASGPAVSPFAREVADLLVLQMKHQHEPQEAASTATAATAAASPNDN 118
DALLA SAL+NASGP+ + FA ++ ++ +++ +S A AAA+ +
Sbjct 687 ITNDALLATSALVNASGPSTAAFAEDIVCIIASGLENSTGSAAGSSGGGAGAAAATSG-S 745
Query 119 STPDELQAARICIELVGDLSRALGTEFG 146
ST DELQ RIC+ELVGD+SRALG F
Sbjct 746 STTDELQTVRICVELVGDVSRALGPAFA 773
> hsa:55565 ZNF821, FLJ16350; zinc finger protein 821
Length=412
Score = 35.8 bits (81), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 42/99 (42%), Gaps = 21/99 (21%)
Query 37 PSQAEGS----TSPNNPQGFVVSSEPSVCDALLALSALINASGPAVSPFAREVADLLVLQ 92
PS AEG + P P G ++ VC+ A L+ A P+V +A
Sbjct 215 PSSAEGKDIAFSPPVYPAGILL-----VCNNCAAYRKLLEAQTPSVRKWA---------- 259
Query 93 MKHQHEPQEAA--STATAATAAASPNDNSTPDELQAARI 129
++ Q+EP E TA S DN TP+E + R+
Sbjct 260 LRRQNEPLEVRLQRLERERTAKKSRRDNETPEEREVRRM 298
> mmu:75871 Zfp821, 4930566A11Rik, Znf821; zinc finger protein
821
Length=413
Score = 34.3 bits (77), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 28/99 (28%), Positives = 41/99 (41%), Gaps = 21/99 (21%)
Query 37 PSQAEGS----TSPNNPQGFVVSSEPSVCDALLALSALINASGPAVSPFAREVADLLVLQ 92
PS AEG + P P G ++ VC+ A L+ P+V +A
Sbjct 216 PSSAEGKDIACSPPVYPAGILL-----VCNNCAAYRKLLETQTPSVRKWA---------- 260
Query 93 MKHQHEPQEAA--STATAATAAASPNDNSTPDELQAARI 129
++ Q+EP E TA S DN TP+E + R+
Sbjct 261 LRRQNEPLEVRLQRLERERTAKKSRRDNETPEEREVRRM 299
> tpv:TP01_0465 histone acetyltransferase Gcn5; K06062 histone
acetyltransferase [EC:2.3.1.48]
Length=631
Score = 29.3 bits (64), Expect = 5.3, Method: Composition-based stats.
Identities = 12/62 (19%), Positives = 33/62 (53%), Gaps = 2/62 (3%)
Query 38 SQAEGSTSPNNPQGFVVSSEPSVCDALLALSALINASGPAVSPFAREVADLLVLQMKHQH 97
+++ S P+ P+ + ++ L +++ +N+ +++ + RE+ +L VL K +
Sbjct 13 NESTSSPGPSTPKS--ATEAKTISRPLASVNTELNSKNESITSYVREIVNLYVLDCKKRP 70
Query 98 EP 99
+P
Sbjct 71 DP 72
Lambda K H
0.313 0.128 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40