bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3251_orf2
Length=73
Score E
Sequences producing significant alignments: (Bits) Value
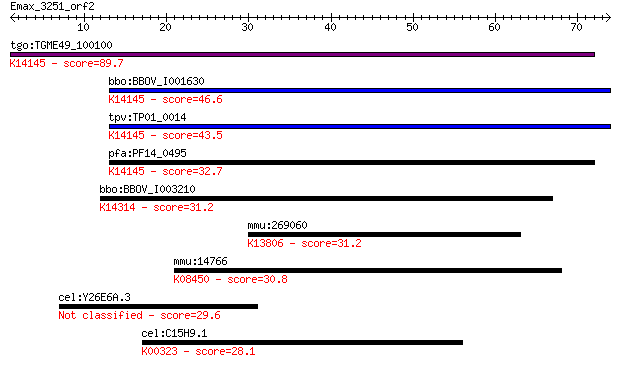
tgo:TGME49_100100 rhoptry neck protein 2 (EC:3.2.1.39); K14145... 89.7 2e-18
bbo:BBOV_I001630 19.m02019; hypothetical protein; K14145 rhopt... 46.6 2e-05
tpv:TP01_0014 hypothetical protein; K14145 rhoptry neck protein 2 43.5
pfa:PF14_0495 PfRON2; rhoptry neck protein 2; K14145 rhoptry n... 32.7 0.26
bbo:BBOV_I003210 19.m02136; hypothetical protein; K14314 nucle... 31.2 0.80
mmu:269060 Dagla, KIAA0659, Nsddr; diacylglycerol lipase, alph... 31.2 0.87
mmu:14766 Gpr56, Cyt28, TM7LN4, TM7XN1; G protein-coupled rece... 30.8 1.1
cel:Y26E6A.3 hypothetical protein 29.6 2.6
cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase fa... 28.1 7.5
> tgo:TGME49_100100 rhoptry neck protein 2 (EC:3.2.1.39); K14145
rhoptry neck protein 2
Length=1404
Score = 89.7 bits (221), Expect = 2e-18, Method: Composition-based stats.
Identities = 40/71 (56%), Positives = 55/71 (77%), Gaps = 0/71 (0%)
Query 1 ISYDKIAIPDLTRWEKEINNKWLHALSAYLSHPYGAAAAAAGDPVALLVKESKERLEKEI 60
+S+ I IP+LT W+ ++N+KWL A +AYL HPYG AA A DPVALLVK+S++RL+ E
Sbjct 858 MSFGGIPIPNLTNWDAQLNSKWLEAYNAYLRHPYGRAALNARDPVALLVKDSRDRLQAEA 917
Query 61 DNTIFLGRVVQ 71
+ TIFLGR+ +
Sbjct 918 EGTIFLGRIAK 928
> bbo:BBOV_I001630 19.m02019; hypothetical protein; K14145 rhoptry
neck protein 2
Length=1365
Score = 46.6 bits (109), Expect = 2e-05, Method: Composition-based stats.
Identities = 23/61 (37%), Positives = 34/61 (55%), Gaps = 0/61 (0%)
Query 13 RWEKEINNKWLHALSAYLSHPYGAAAAAAGDPVALLVKESKERLEKEIDNTIFLGRVVQT 72
RWEK +N+ L +A+L P + +VK+S++ LE+ +D TIF GRVV
Sbjct 867 RWEKHLNSLILDGYNAFLDLPSIKVLDGRHSLIYEIVKDSRDNLEQHLDETIFFGRVVNP 926
Query 73 P 73
P
Sbjct 927 P 927
> tpv:TP01_0014 hypothetical protein; K14145 rhoptry neck protein
2
Length=788
Score = 43.5 bits (101), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 39/63 (61%), Gaps = 4/63 (6%)
Query 13 RWEKEINNKWLHALSAYLSHPYGAAAAAAGDPVAL--LVKESKERLEKEIDNTIFLGRVV 70
+W+K +N + L + + +L G+ A G L +VK+S++ LE+ I+NTIF GR++
Sbjct 298 KWDKSLNTEILKSFNEFLE--LGSVKALEGRNHILYEVVKDSRDNLEQNIENTIFFGRIL 355
Query 71 QTP 73
+P
Sbjct 356 NSP 358
> pfa:PF14_0495 PfRON2; rhoptry neck protein 2; K14145 rhoptry
neck protein 2
Length=2189
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Query 13 RWEKEINNKWLHALSAYLSHPYGAAAAAAGDPVALLVKESKERLEKEIDNTIFLGRVVQ 71
+W++ IN + + AL +L + A V +V++SK LE IDN I R V+
Sbjct 1688 KWDQFINKELVRALGLWLE--FNDNPTNASSFVYKVVEDSKHLLENNIDNNIIFSRTVK 1744
> bbo:BBOV_I003210 19.m02136; hypothetical protein; K14314 nuclear
pore complex protein Nup210
Length=2236
Score = 31.2 bits (69), Expect = 0.80, Method: Composition-based stats.
Identities = 18/61 (29%), Positives = 29/61 (47%), Gaps = 6/61 (9%)
Query 12 TRWEKEINNKWLHALSAYLSHPYGAAAAAAGD------PVALLVKESKERLEKEIDNTIF 65
T+W+ +N HA A + P+G + PVA L KES ++ID +I+
Sbjct 907 TKWQVRVNQFGTHAFRAVVYDPFGIPLCPIHNYNLSWRPVAQLPKESGTASYQKIDGSIY 966
Query 66 L 66
+
Sbjct 967 I 967
> mmu:269060 Dagla, KIAA0659, Nsddr; diacylglycerol lipase, alpha;
K13806 sn1-specific diacylglycerol lipase alpha [EC:3.1.1.-]
Length=1044
Score = 31.2 bits (69), Expect = 0.87, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 30 LSHPYGAAAAAAGDPVALLVKESKERLEKEIDN 62
L P AA+AA DPV LL+ ++ERL E+ +
Sbjct 742 LLSPVAAASAARQDPVELLLLSTQERLAAELQS 774
> mmu:14766 Gpr56, Cyt28, TM7LN4, TM7XN1; G protein-coupled receptor
56; K08450 G protein-coupled receptor 56
Length=687
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 23/47 (48%), Gaps = 4/47 (8%)
Query 21 KWLHALSAYLSHPYGAAAAAAGDPVALLVKESKERLEKEIDNTIFLG 67
K L LS YL HP AA P A + + + LE ++ + FLG
Sbjct 181 KELQQLSRYLQHP----QKAAKRPTAAFISQQLQSLESKLTSVSFLG 223
> cel:Y26E6A.3 hypothetical protein
Length=204
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 10/24 (41%), Positives = 18/24 (75%), Gaps = 0/24 (0%)
Query 7 AIPDLTRWEKEINNKWLHALSAYL 30
A+PD+T E+N ++LH++ +YL
Sbjct 45 ALPDVTFQPTEVNGRYLHSIVSYL 68
> cel:C15H9.1 nnt-1; Nicotinamide Nucleotide Transhydrogenase
family member (nnt-1); K00323 NAD(P) transhydrogenase [EC:1.6.1.2]
Length=1041
Score = 28.1 bits (61), Expect = 7.5, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 17 EINNKWLHALSAYLSHPYGAAAAAAGDPVALLVKESKER 55
E + L+A S + YG AA A P+A LVKE ++R
Sbjct 872 ETADMLLNARSVIIIPGYGLCAAQAQYPIAQLVKELQQR 910
Lambda K H
0.315 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2011623444
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40