bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
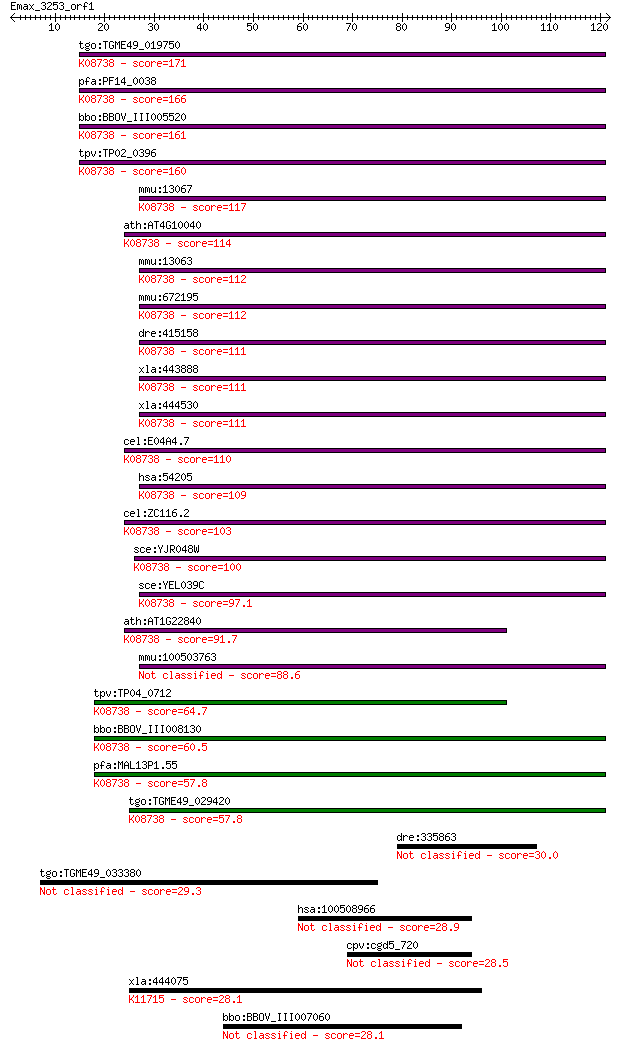
Query= Emax_3253_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019750 cytochrome c, putative ; K08738 cytochrome c 171 6e-43
pfa:PF14_0038 cytochrome c, putative; K08738 cytochrome c 166 2e-41
bbo:BBOV_III005520 17.m07493; cytochrome c; K08738 cytochrome c 161 4e-40
tpv:TP02_0396 cytochrome c; K08738 cytochrome c 160 8e-40
mmu:13067 Cyct, T-Cc; cytochrome c, testis; K08738 cytochrome c 117 8e-27
ath:AT4G10040 CYTC-2; CYTC-2 (cytochrome c-2); electron carrie... 114 5e-26
mmu:13063 Cycs; cytochrome c, somatic; K08738 cytochrome c 112 2e-25
mmu:672195 Gm10053; predicted gene 10053; K08738 cytochrome c 112 2e-25
dre:415158 cycsb, cyc, wu:fk52b01, zgc:86706; cytochrome c, so... 111 5e-25
xla:443888 cyct-a, MGC80124, cycs-a; cytochrome c, testis; K08... 111 5e-25
xla:444530 cycs, cyct; cytochrome c, somatic; K08738 cytochrome c 111 5e-25
cel:E04A4.7 cyc-2.1; CYtochrome C family member (cyc-2.1); K08... 110 1e-24
hsa:54205 CYCS, CYC, HCS, THC4; cytochrome c, somatic; K08738 ... 109 2e-24
cel:ZC116.2 cyc-2.2; CYtochrome C family member (cyc-2.2); K08... 103 1e-22
sce:YJR048W CYC1; Cyc1p; K08738 cytochrome c 100 1e-21
sce:YEL039C CYC7; Cyc7p; K08738 cytochrome c 97.1 1e-20
ath:AT1G22840 CYTC-1; CYTC-1 (CYTOCHROME C-1); electron carrie... 91.7 6e-19
mmu:100503763 cytochrome c, somatic-like 88.6 4e-18
tpv:TP04_0712 cytochrome c; K08738 cytochrome c 64.7 7e-11
bbo:BBOV_III008130 17.m07711; cytochrome c family protein; K08... 60.5 1e-09
pfa:MAL13P1.55 cytochrome c2 precursor,putative; K08738 cytoch... 57.8 8e-09
tgo:TGME49_029420 cytochrome c, putative ; K08738 cytochrome c 57.8 9e-09
dre:335863 itm2cb, fj35c02, itm2c, wu:fj35c02, zgc:66349; inte... 30.0 1.9
tgo:TGME49_033380 hypothetical protein 29.3 2.9
hsa:100508966 putative uncharacterized protein ENSP00000334305... 28.9 4.7
cpv:cgd5_720 choline/ethanolamine kinase family protein 28.5 5.8
xla:444075 ern2, Ire1beta, MGC83537, ire1b; endoplasmic reticu... 28.1 6.4
bbo:BBOV_III007060 17.m10570; membrane protein 28.1 7.3
> tgo:TGME49_019750 cytochrome c, putative ; K08738 cytochrome
c
Length=115
Score = 171 bits (432), Expect = 6e-43, Method: Compositional matrix adjust.
Identities = 78/106 (73%), Positives = 86/106 (81%), Gaps = 0/106 (0%)
Query 15 MSRPDPDVTVPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSADFPYS 74
MSR +PDV VP+GD KAGAKVFKAKC+QCHT+N GGAAK GPNL GF+GR SG ADFPYS
Sbjct 1 MSRAEPDVQVPSGDIKAGAKVFKAKCSQCHTINAGGAAKAGPNLHGFIGRESGKADFPYS 60
Query 75 EANKNSGIVWSEKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
+ANK SGI+WSEKHLWEYL+NPK YIPGTKMVFA R I
Sbjct 61 DANKTSGIIWSEKHLWEYLLNPKTYIPGTKMVFAGIKKEKERADLI 106
> pfa:PF14_0038 cytochrome c, putative; K08738 cytochrome c
Length=115
Score = 166 bits (420), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 75/106 (70%), Positives = 85/106 (80%), Gaps = 0/106 (0%)
Query 15 MSRPDPDVTVPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSADFPYS 74
MSRP+P V VP GD K GAK+FKAKCAQCHT+NKGGA KQGPNL GF GR SG +DFPYS
Sbjct 1 MSRPEPQVQVPEGDYKKGAKLFKAKCAQCHTINKGGAVKQGPNLHGFYGRKSGDSDFPYS 60
Query 75 EANKNSGIVWSEKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
+ANKNSGI+WS+KHL+EYL+NPK YIPGTKM+FA R I
Sbjct 61 DANKNSGIIWSDKHLFEYLLNPKLYIPGTKMIFAGIKKEKERADLI 106
> bbo:BBOV_III005520 17.m07493; cytochrome c; K08738 cytochrome
c
Length=115
Score = 161 bits (408), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 72/106 (67%), Positives = 86/106 (81%), Gaps = 0/106 (0%)
Query 15 MSRPDPDVTVPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSADFPYS 74
MS+P+P+VTVP GD + GAK+FK+KCAQCHT NKGGA KQGPNL+G GRASGSAD+ Y+
Sbjct 1 MSKPEPNVTVPDGDAQKGAKLFKSKCAQCHTTNKGGATKQGPNLYGLFGRASGSADYAYT 60
Query 75 EANKNSGIVWSEKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
+ANK+SGIVWS+KHL+ YLVNPK YIPGTKM+FA R I
Sbjct 61 DANKSSGIVWSDKHLFVYLVNPKEYIPGTKMIFAGLKKEQDRADII 106
> tpv:TP02_0396 cytochrome c; K08738 cytochrome c
Length=115
Score = 160 bits (405), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 70/106 (66%), Positives = 85/106 (80%), Gaps = 0/106 (0%)
Query 15 MSRPDPDVTVPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSADFPYS 74
M++P+PDV +P GD GAK+FK+KCAQCHT+NKGG+ KQGPNL+GF GR SG++D+ YS
Sbjct 1 MAKPEPDVVIPEGDSSKGAKLFKSKCAQCHTINKGGSVKQGPNLYGFYGRKSGASDYAYS 60
Query 75 EANKNSGIVWSEKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
+ANKNSGIVWS+KHL+ YLVNPK YIPGTKMVFA R I
Sbjct 61 DANKNSGIVWSDKHLFVYLVNPKQYIPGTKMVFAGLKKEQDRADLI 106
> mmu:13067 Cyct, T-Cc; cytochrome c, testis; K08738 cytochrome
c
Length=105
Score = 117 bits (293), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 55/95 (57%), Positives = 65/95 (68%), Gaps = 1/95 (1%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSA-DFPYSEANKNSGIVWS 85
GD +AG K+F KCAQCHTV KGG K GPNL+G GR +G A F Y++ANKN G++WS
Sbjct 2 GDAEAGKKIFVQKCAQCHTVEKGGKHKTGPNLWGLFGRKTGQAPGFSYTDANKNKGVIWS 61
Query 86 EKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E+ L EYL NPK YIPGTKM+FA R I
Sbjct 62 EETLMEYLENPKKYIPGTKMIFAGIKKKSEREDLI 96
> ath:AT4G10040 CYTC-2; CYTC-2 (cytochrome c-2); electron carrier/
heme binding / iron ion binding; K08738 cytochrome c
Length=112
Score = 114 bits (286), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 54/98 (55%), Positives = 64/98 (65%), Gaps = 1/98 (1%)
Query 24 VPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSA-DFPYSEANKNSGI 82
P G+PKAG K+F+ KCAQCHTV KG KQGPNL G GR SG+ + YS ANK+ +
Sbjct 7 APPGNPKAGEKIFRTKCAQCHTVEKGAGHKQGPNLNGLFGRQSGTTPGYSYSAANKSMAV 66
Query 83 VWSEKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
W EK L++YL+NPK YIPGTKMVF R I
Sbjct 67 NWEEKTLYDYLLNPKKYIPGTKMVFPGLKKPQDRADLI 104
> mmu:13063 Cycs; cytochrome c, somatic; K08738 cytochrome c
Length=105
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/95 (56%), Positives = 61/95 (64%), Gaps = 1/95 (1%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASG-SADFPYSEANKNSGIVWS 85
GD + G K+F KCAQCHTV KGG K GPNL G GR +G +A F Y++ANKN GI W
Sbjct 2 GDVEKGKKIFVQKCAQCHTVEKGGKHKTGPNLHGLFGRKTGQAAGFSYTDANKNKGITWG 61
Query 86 EKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E L EYL NPK YIPGTKM+FA R I
Sbjct 62 EDTLMEYLENPKKYIPGTKMIFAGIKKKGERADLI 96
> mmu:672195 Gm10053; predicted gene 10053; K08738 cytochrome
c
Length=105
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 54/95 (56%), Positives = 61/95 (64%), Gaps = 1/95 (1%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASG-SADFPYSEANKNSGIVWS 85
GD + G K+F KCAQCHTV KGG K GPNL G GR +G +A F Y++ANKN GI W
Sbjct 2 GDVEKGKKIFVQKCAQCHTVEKGGKHKTGPNLHGLFGRKTGQAAGFSYTDANKNKGITWG 61
Query 86 EKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E L EYL NPK YIPGTKM+FA R I
Sbjct 62 EDTLMEYLENPKKYIPGTKMIFAGIKKKGERADLI 96
> dre:415158 cycsb, cyc, wu:fk52b01, zgc:86706; cytochrome c,
somatic b; K08738 cytochrome c
Length=104
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/95 (56%), Positives = 62/95 (65%), Gaps = 1/95 (1%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSAD-FPYSEANKNSGIVWS 85
GD + G KVF KCAQCHTV GG K GPNL+G GR +G A+ F Y++ANK+ GIVW
Sbjct 2 GDVEKGKKVFVQKCAQCHTVENGGKHKVGPNLWGLFGRKTGQAEGFSYTDANKSKGIVWG 61
Query 86 EKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E L EYL NPK YIPGTKM+FA R I
Sbjct 62 EDTLMEYLENPKKYIPGTKMIFAGIKKKGERADLI 96
> xla:443888 cyct-a, MGC80124, cycs-a; cytochrome c, testis; K08738
cytochrome c
Length=105
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 53/95 (55%), Positives = 63/95 (66%), Gaps = 1/95 (1%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSA-DFPYSEANKNSGIVWS 85
GD + G K+F KCAQCHTV K G K GPNL+G GR +G A F Y++ANK+ GIVW
Sbjct 2 GDAEKGKKIFIQKCAQCHTVEKTGKHKTGPNLWGLFGRKTGQAPGFSYTDANKSKGIVWG 61
Query 86 EKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E L+EYL NPK YIPGTKM+FA + R I
Sbjct 62 EDTLFEYLENPKKYIPGTKMIFAGIKKKNERLDLI 96
> xla:444530 cycs, cyct; cytochrome c, somatic; K08738 cytochrome
c
Length=105
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/95 (56%), Positives = 61/95 (64%), Gaps = 1/95 (1%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSAD-FPYSEANKNSGIVWS 85
GD + G KVF KC+QCHTV KGG K GPNL G GR +G A+ F Y++ANKN GIVW
Sbjct 2 GDVEKGKKVFVQKCSQCHTVEKGGKHKTGPNLHGLFGRKTGQAEGFSYTDANKNKGIVWD 61
Query 86 EKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E L YL NPK YIPGTKM+FA R I
Sbjct 62 EDTLMVYLENPKKYIPGTKMIFAGIKKKGERQDLI 96
> cel:E04A4.7 cyc-2.1; CYtochrome C family member (cyc-2.1); K08738
cytochrome c
Length=111
Score = 110 bits (275), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 52/98 (53%), Positives = 65/98 (66%), Gaps = 2/98 (2%)
Query 24 VPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSAD-FPYSEANKNSGI 82
+PAGD + G KV+K +C QCH V+ A K GP L G +GR SG+ F YS ANKN G+
Sbjct 4 IPAGDYEKGKKVYKQRCLQCHVVD-STATKTGPTLHGVIGRTSGTVSGFDYSAANKNKGV 62
Query 83 VWSEKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
VW+ + L+EYL+NPK YIPGTKMVFA + R I
Sbjct 63 VWTRETLFEYLLNPKKYIPGTKMVFAGLKKADERADLI 100
> hsa:54205 CYCS, CYC, HCS, THC4; cytochrome c, somatic; K08738
cytochrome c
Length=105
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 51/95 (53%), Positives = 59/95 (62%), Gaps = 1/95 (1%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSA-DFPYSEANKNSGIVWS 85
GD + G K+F KC+QCHTV KGG K GPNL G GR +G A + Y+ ANKN GI+W
Sbjct 2 GDVEKGKKIFIMKCSQCHTVEKGGKHKTGPNLHGLFGRKTGQAPGYSYTAANKNKGIIWG 61
Query 86 EKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E L EYL NPK YIPGTKM+F R I
Sbjct 62 EDTLMEYLENPKKYIPGTKMIFVGIKKKEERADLI 96
> cel:ZC116.2 cyc-2.2; CYtochrome C family member (cyc-2.2); K08738
cytochrome c
Length=123
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 50/98 (51%), Positives = 61/98 (62%), Gaps = 2/98 (2%)
Query 24 VPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGS-ADFPYSEANKNSGI 82
+P GD + G K+FK +C QCH VN K GP L G +GR SG A F YS ANKN G+
Sbjct 14 IPEGDNEKGKKIFKQRCEQCHVVN-SLQTKTGPTLNGVIGRQSGQVAGFDYSAANKNKGV 72
Query 83 VWSEKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
VW + L++YL +PK YIPGTKMVFA + R I
Sbjct 73 VWDRQTLFDYLADPKKYIPGTKMVFAGLKKADERADLI 110
> sce:YJR048W CYC1; Cyc1p; K08738 cytochrome c
Length=109
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 48/96 (50%), Positives = 58/96 (60%), Gaps = 1/96 (1%)
Query 26 AGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSAD-FPYSEANKNSGIVW 84
AG K GA +FK +C QCHTV KGG K GPNL G GR SG A+ + Y++AN ++W
Sbjct 6 AGSAKKGATLFKTRCLQCHTVEKGGPHKVGPNLHGIFGRHSGQAEGYSYTDANIKKNVLW 65
Query 85 SEKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E ++ EYL NPK YIPGTKM F R I
Sbjct 66 DENNMSEYLTNPKKYIPGTKMAFGGLKKEKDRNDLI 101
> sce:YEL039C CYC7; Cyc7p; K08738 cytochrome c
Length=113
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 46/95 (48%), Positives = 55/95 (57%), Gaps = 1/95 (1%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSAD-FPYSEANKNSGIVWS 85
G K GA +FK +C QCHT+ +GG K GPNL G GR SG + Y++AN N + W
Sbjct 11 GSAKKGATLFKTRCQQCHTIEEGGPNKVGPNLHGIFGRHSGQVKGYSYTDANINKNVKWD 70
Query 86 EKHLWEYLVNPKAYIPGTKMVFASFDNSHRRCSRI 120
E + EYL NPK YIPGTKM FA R I
Sbjct 71 EDSMSEYLTNPKKYIPGTKMAFAGLKKEKDRNDLI 105
> ath:AT1G22840 CYTC-1; CYTC-1 (CYTOCHROME C-1); electron carrier/
heme binding / iron ion binding; K08738 cytochrome c
Length=102
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 43/78 (55%), Positives = 53/78 (67%), Gaps = 1/78 (1%)
Query 24 VPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASG-SADFPYSEANKNSGI 82
P G+ KAG K+F+ KCAQCHTV G KQGPNL G GR SG +A + YS ANKN +
Sbjct 7 APPGNAKAGEKIFRTKCAQCHTVEAGAGHKQGPNLNGLFGRQSGTTAGYSYSAANKNKAV 66
Query 83 VWSEKHLWEYLVNPKAYI 100
W EK L++YL+NPK +
Sbjct 67 EWEEKALYDYLLNPKKVL 84
> mmu:100503763 cytochrome c, somatic-like
Length=106
Score = 88.6 bits (218), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 44/96 (45%), Positives = 59/96 (61%), Gaps = 2/96 (2%)
Query 27 GDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASG-SADFPYSEANKNSGIVWS 85
GD + G K+F +CAQCHTV KGG K GPNL G G+ +G +A F Y++A+KN GI W
Sbjct 2 GDVEKGKKIFVQRCAQCHTVRKGGKHKTGPNLQGLFGQKTGQAAGFSYTDASKNRGITWG 61
Query 86 EKHLWEYLV-NPKAYIPGTKMVFASFDNSHRRCSRI 120
E L + + + YIPGTKM+FA + + I
Sbjct 62 EDTLLDGVFRESQKYIPGTKMIFAGIKKTGEKKDLI 97
> tpv:TP04_0712 cytochrome c; K08738 cytochrome c
Length=145
Score = 64.7 bits (156), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 34/92 (36%), Positives = 50/92 (54%), Gaps = 10/92 (10%)
Query 18 PDPDVTVPAGDPKAGAKVFKAKCAQCHTV-----NKGGAAKQGPNLFGFLGRASGSADF- 71
PD D +P G + GAK+FK C QCH++ GG + GP LF GR SG +
Sbjct 11 PD-DFVLPEGSAERGAKLFKKHCKQCHSMRPDNRQTGGFSSIGPTLFNVYGRTSGIQNLG 69
Query 72 ---PYSEANKNSGIVWSEKHLWEYLVNPKAYI 100
+ + K+SGIVW + +L Y+ NP+ ++
Sbjct 70 GLNLMTHSMKSSGIVWDDANLMRYMKNPQLFV 101
> bbo:BBOV_III008130 17.m07711; cytochrome c family protein; K08738
cytochrome c
Length=146
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/113 (31%), Positives = 58/113 (51%), Gaps = 11/113 (9%)
Query 18 PDPDVTVPAGDPKAGAKVFKAKCAQCHTV-----NKGGAAKQGPNLFGFLGRASGSADFP 72
PD + +PAG+ + GAK+FK C QCH++ G A GP LF R +G++
Sbjct 11 PD-NFVLPAGNAQRGAKLFKKHCQQCHSMRPDNRQTSGFATVGPTLFNVYCRTAGASGHD 69
Query 73 ----YSEANKNSGIVWSEKHLWEYLVNPKAYIPG-TKMVFASFDNSHRRCSRI 120
++ +N+GIVW++ +L Y+ NP+ Y+ M F+ N R +
Sbjct 70 SVKGITDNLQNAGIVWTDANLMRYMKNPERYVNAVVGMNFSGLPNFQDRVDIV 122
> pfa:MAL13P1.55 cytochrome c2 precursor,putative; K08738 cytochrome
c
Length=159
Score = 57.8 bits (138), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 35/114 (30%), Positives = 52/114 (45%), Gaps = 12/114 (10%)
Query 18 PDPDVTVPAGDPKAGAKVFKAKCAQCHTV------NKGGAAKQGPNLFGFLGRAS----G 67
PD D +P GD G K+FK C QCH++ G GP LF R + G
Sbjct 17 PD-DFVLPPGDKVKGEKLFKKHCKQCHSIAPDNSQTNSGFTSWGPTLFNVYNRTAGMSKG 75
Query 68 SADFPYSEANKNSGIVWSEKHLWEYLVNPKAYIPG-TKMVFASFDNSHRRCSRI 120
++ F S SGI+W++ +L +Y+ NP+ ++ M F N R +
Sbjct 76 NSPFQTSPDLYTSGIIWNDVNLLKYMKNPQQFVESHIGMNFKGLSNLQERVDIV 129
> tgo:TGME49_029420 cytochrome c, putative ; K08738 cytochrome
c
Length=163
Score = 57.8 bits (138), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 38/107 (35%), Positives = 51/107 (47%), Gaps = 11/107 (10%)
Query 25 PAGDPKAGAKVFKAKCAQCHTVNK------GGAAKQGPNLFGFLGRASG----SADFPYS 74
P GD GAK+FK CAQCH++ G GP L+ R +G S+ P S
Sbjct 31 PPGDAVRGAKLFKKHCAQCHSIFPDGRHLIAGNTSWGPTLWNVYMRTAGVEKDSSCSPIS 90
Query 75 EANKNSGIVWSEKHLWEYLVNPKAYIPG-TKMVFASFDNSHRRCSRI 120
+SG+VW++ +L Y+ NPK +I G M F N R I
Sbjct 91 SHILDSGVVWNDANLMRYMKNPKMFINGVVGMNFFGIANFQDRVDII 137
> dre:335863 itm2cb, fj35c02, itm2c, wu:fj35c02, zgc:66349; integral
membrane protein 2Cb
Length=260
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 2/30 (6%)
Query 79 NSGIVWSEKHLWEYLVNPK--AYIPGTKMV 106
N+ IV ++LWE LVN K Y+P T M+
Sbjct 160 NTTIVMPPRNLWELLVNVKRGTYLPQTYMI 189
> tgo:TGME49_033380 hypothetical protein
Length=1049
Score = 29.3 bits (64), Expect = 2.9, Method: Composition-based stats.
Identities = 21/71 (29%), Positives = 27/71 (38%), Gaps = 10/71 (14%)
Query 7 CGLIIFHKMSRPDPDVTVPAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGP---NLFGFLG 63
C + M+ PDV PA DP G +VF+ + G K P G L
Sbjct 655 CSSFVVPPMTSATPDVAEPADDPHLGQQVFRP-------IENGRYLKNTPPPAPFHGSLR 707
Query 64 RASGSADFPYS 74
S FPY+
Sbjct 708 AGSFVGHFPYA 718
> hsa:100508966 putative uncharacterized protein ENSP00000334305-like
Length=771
Score = 28.9 bits (63), Expect = 4.7, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 24/37 (64%), Gaps = 4/37 (10%)
Query 59 FGFLGRASGSADFPYSEANK--NSGIVWSEKHLWEYL 93
+G LG+A GSA P++ A K ++GI E+H WE +
Sbjct 737 YGVLGQAQGSA--PWTSAFKTFSAGIAGLERHAWETM 771
> cpv:cgd5_720 choline/ethanolamine kinase family protein
Length=443
Score = 28.5 bits (62), Expect = 5.8, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 14/25 (56%), Gaps = 0/25 (0%)
Query 69 ADFPYSEANKNSGIVWSEKHLWEYL 93
D PYSE+NK+ I+W W L
Sbjct 175 GDVPYSESNKSISILWPTLDKWASL 199
> xla:444075 ern2, Ire1beta, MGC83537, ire1b; endoplasmic reticulum
to nucleus signaling 2 (EC:2.7.11.1); K11715 serine/threonine-protein
kinase/endoribonuclease IRE2 [EC:2.7.11.1 3.1.26.-]
Length=958
Score = 28.1 bits (61), Expect = 6.4, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 32/71 (45%), Gaps = 3/71 (4%)
Query 25 PAGDPKAGAKVFKAKCAQCHTVNKGGAAKQGPNLFGFLGRASGSADFPYSEANKNSGIVW 84
P +P A +F A C + ++K G G NL SGS P + + + +V
Sbjct 738 PKQNPTAAVDIFSAGCVFYYVLSK-GQHPFGDNLRRQSNILSGSYSLPKLQDDTHENVV- 795
Query 85 SEKHLWEYLVN 95
+HL E ++N
Sbjct 796 -ARHLVEMMIN 805
> bbo:BBOV_III007060 17.m10570; membrane protein
Length=841
Score = 28.1 bits (61), Expect = 7.3, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 25/50 (50%), Gaps = 2/50 (4%)
Query 44 HTVNKGGAAKQGPNLFGFLGRASGSADFPYSEANKNSGIVWSEK--HLWE 91
H+ GG + PNLF S D +EA+ NS + +E+ H+W+
Sbjct 255 HSDMNGGITEISPNLFSDKTERQTSTDNDTNEASGNSYVYSAEELLHIWD 304
Lambda K H
0.321 0.136 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2013067560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40