bitscore colors: <40, 40-50 , 50-80, 80-200, >200
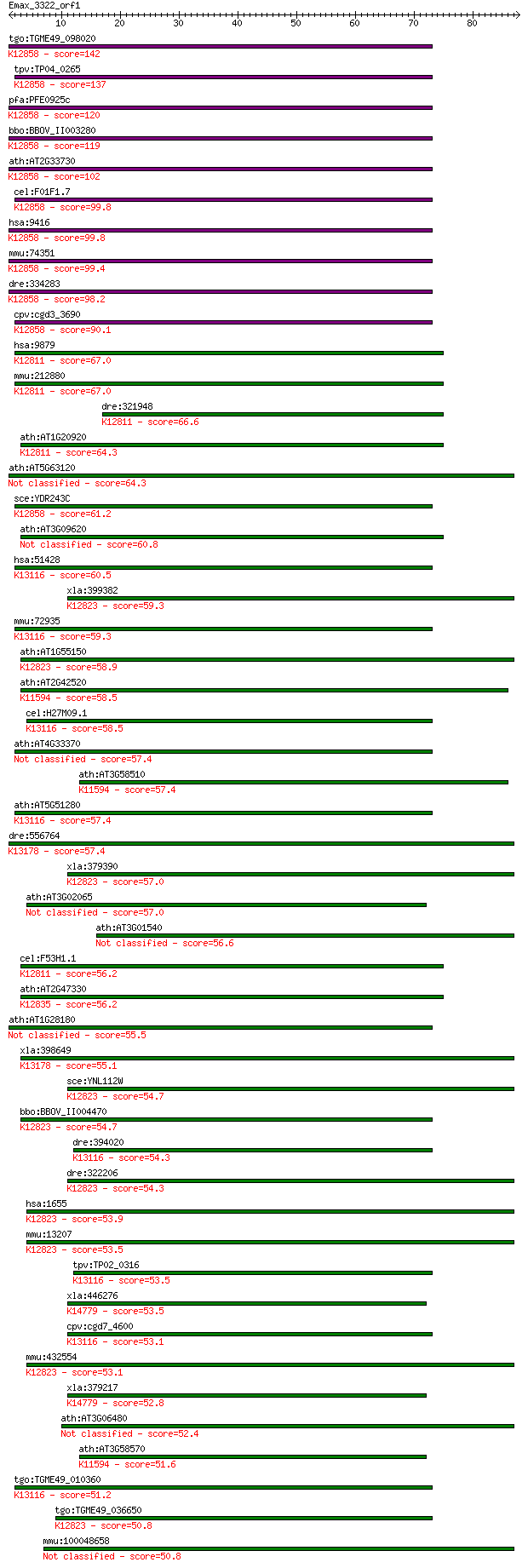
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3322_orf1
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putativ... 142 3e-34
tpv:TP04_0265 small nuclear ribonucleoprotein; K12858 ATP-depe... 137 6e-33
pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA... 120 1e-27
bbo:BBOV_II003280 18.m06276; DEAD box RNA helicase; K12858 ATP... 119 2e-27
ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-depe... 102 3e-22
cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member (d... 99.8 2e-21
hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEA... 99.8 2e-21
mmu:74351 Ddx23, 3110082M05Rik, 4921506D17Rik; DEAD (Asp-Glu-A... 99.4 2e-21
dre:334283 ddx23, wu:fi39b12, zgc:63742; DEAD (Asp-Glu-Ala-Asp... 98.2 5e-21
cpv:cgd3_3690 U5 snRNP 100 kD protein ; K12858 ATP-dependent R... 90.1 2e-18
hsa:9879 DDX46, FLJ25329, KIAA0801, MGC9936, PRPF5, Prp5; DEAD... 67.0 1e-11
mmu:212880 Ddx46, 2200005K02Rik, 8430438J23Rik, AI325430, AI95... 67.0 2e-11
dre:321948 ddx46, fb39a03, wu:fb39a03; DEAD (Asp-Glu-Ala-Asp) ... 66.6 2e-11
ath:AT1G20920 DEAD box RNA helicase, putative; K12811 ATP-depe... 64.3 8e-11
ath:AT5G63120 ethylene-responsive DEAD box RNA helicase, putat... 64.3 1e-10
sce:YDR243C PRP28; Prp28p (EC:3.6.1.-); K12858 ATP-dependent R... 61.2 7e-10
ath:AT3G09620 DEAD/DEAH box helicase, putative 60.8 1e-09
hsa:51428 DDX41, ABS, MGC8828; DEAD (Asp-Glu-Ala-Asp) box poly... 60.5 1e-09
xla:399382 ddx5, MGC81559; DEAD (Asp-Glu-Ala-Asp) box polypept... 59.3 3e-09
mmu:72935 Ddx41, 2900024F02Rik, AA958953, ABS, AI324246; DEAD ... 59.3 3e-09
ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823 A... 58.9 4e-09
ath:AT2G42520 DEAD box RNA helicase, putative; K11594 ATP-depe... 58.5 5e-09
cel:H27M09.1 hypothetical protein; K13116 ATP-dependent RNA he... 58.5 5e-09
ath:AT4G33370 DEAD-box protein abstrakt, putative 57.4 1e-08
ath:AT3G58510 DEAD box RNA helicase, putative (RH11); K11594 A... 57.4 1e-08
ath:AT5G51280 DEAD-box protein abstrakt, putative; K13116 ATP-... 57.4 1e-08
dre:556764 similar to Probable RNA-dependent helicase p72 (DEA... 57.4 1e-08
xla:379390 MGC53795; similar to DEAD/H (Asp-Glu-Ala-Asp/His) b... 57.0 1e-08
ath:AT3G02065 DEAD/DEAH box helicase family protein 57.0 1e-08
ath:AT3G01540 DRH1; DRH1 (DEAD BOX RNA HELICASE 1); ATP-depend... 56.6 2e-08
cel:F53H1.1 hypothetical protein; K12811 ATP-dependent RNA hel... 56.2 2e-08
ath:AT2G47330 DEAD/DEAH box helicase, putative; K12835 ATP-dep... 56.2 2e-08
ath:AT1G28180 ATP binding / ATP-dependent helicase/ helicase/ ... 55.5 4e-08
xla:398649 ddx17, MGC80019; DEAD (Asp-Glu-Ala-Asp) box polypep... 55.1 5e-08
sce:YNL112W DBP2; Dbp2p (EC:3.6.1.-); K12823 ATP-dependent RNA... 54.7 6e-08
bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-depe... 54.7 6e-08
dre:394020 ddx41, MGC55896, wu:fb92e02, zgc:55896; DEAD (Asp-G... 54.3 9e-08
dre:322206 ddx5, wu:fa56a07, wu:fb11e01, wu:fb16c10, wu:fb53b0... 54.3 1e-07
hsa:1655 DDX5, DKFZp434E109, DKFZp686J01190, G17P1, HLR1, HUMP... 53.9 1e-07
mmu:13207 Ddx5, 2600009A06Rik, G17P1, HUMP68, Hlr1, MGC118083,... 53.5 1e-07
tpv:TP02_0316 RNA helicase-1; K13116 ATP-dependent RNA helicas... 53.5 2e-07
xla:446276 ddx52; DEAD (Asp-Glu-Ala-Asp) box polypeptide 52; K... 53.5 2e-07
cpv:cgd7_4600 abstrakt protein SF II helicase + Znknuckle C2HC... 53.1 2e-07
mmu:432554 Gm12183, OTTMUSG00000005521; predicted gene 12183; ... 53.1 2e-07
xla:379217 MGC53409; similar to ATP-dependent, RNA helicase; K... 52.8 3e-07
ath:AT3G06480 DEAD box RNA helicase, putative 52.4 3e-07
ath:AT3G58570 DEAD box RNA helicase, putative; K11594 ATP-depe... 51.6 6e-07
tgo:TGME49_010360 DEAD/DEAH box helicase, putative (EC:5.99.1.... 51.2 8e-07
tgo:TGME49_036650 DEAD/DEAH box helicase, putative (EC:5.99.1.... 50.8 9e-07
mmu:100048658 Ddx43, OTTMUSG00000019690; DEAD (Asp-Glu-Ala-Asp... 50.8 1e-06
> tgo:TGME49_098020 DEAD-box ATP-dependent RNA helicase, putative
(EC:2.7.11.25); K12858 ATP-dependent RNA helicase DDX23/PRP28
[EC:3.6.4.13]
Length=1158
Score = 142 bits (357), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 63/72 (87%), Positives = 68/72 (94%), Gaps = 0/72 (0%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
EEM ERDWRIFREDFEIY+KGGRVPPPIRTWAE+ LPWELIEA+K ANY+RPTPIQMQAI
Sbjct 709 EEMNERDWRIFREDFEIYIKGGRVPPPIRTWAESALPWELIEAVKHANYDRPTPIQMQAI 768
Query 61 PIALEMRDLIGM 72
PIALE RDLIG+
Sbjct 769 PIALEQRDLIGI 780
> tpv:TP04_0265 small nuclear ribonucleoprotein; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=744
Score = 137 bits (346), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 62/71 (87%), Positives = 68/71 (95%), Gaps = 0/71 (0%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
EMTERDWRIFREDFEIY+KGGRVPPPIRTWAE+ LPWEL+EAIK+A Y +PTPIQMQAIP
Sbjct 302 EMTERDWRIFREDFEIYIKGGRVPPPIRTWAESPLPWELLEAIKKAGYIKPTPIQMQAIP 361
Query 62 IALEMRDLIGM 72
IALEMRDLIG+
Sbjct 362 IALEMRDLIGI 372
> pfa:PFE0925c snrnp protein, putative; K12858 ATP-dependent RNA
helicase DDX23/PRP28 [EC:3.6.4.13]
Length=1123
Score = 120 bits (300), Expect = 1e-27, Method: Composition-based stats.
Identities = 55/72 (76%), Positives = 64/72 (88%), Gaps = 0/72 (0%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
EEMT+RDWRIFRED EIY+KGG VPPPIR W E+ L +L++AIK+A YE+PTPIQMQAI
Sbjct 670 EEMTDRDWRIFREDNEIYIKGGVVPPPIRKWEESNLSNDLLKAIKKAKYEKPTPIQMQAI 729
Query 61 PIALEMRDLIGM 72
PIALEMRDLIG+
Sbjct 730 PIALEMRDLIGI 741
> bbo:BBOV_II003280 18.m06276; DEAD box RNA helicase; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=714
Score = 119 bits (299), Expect = 2e-27, Method: Composition-based stats.
Identities = 53/72 (73%), Positives = 62/72 (86%), Gaps = 0/72 (0%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
E MT+RDWRIFREDF+IY+KG RVPPP+RTWAE+ LP EL+ AIK A ++ PTPIQMQAI
Sbjct 269 ENMTQRDWRIFREDFDIYVKGTRVPPPMRTWAESNLPSELLRAIKDAGFKSPTPIQMQAI 328
Query 61 PIALEMRDLIGM 72
PI L MRDLIG+
Sbjct 329 PIGLGMRDLIGL 340
> ath:AT2G33730 DEAD box RNA helicase, putative; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=733
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 43/72 (59%), Positives = 60/72 (83%), Gaps = 0/72 (0%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
EEMTERDWRIFREDF I KG R+P P+R+W E++L EL++A+++A Y++P+PIQM AI
Sbjct 285 EEMTERDWRIFREDFNISYKGSRIPRPMRSWEESKLTSELLKAVERAGYKKPSPIQMAAI 344
Query 61 PIALEMRDLIGM 72
P+ L+ RD+IG+
Sbjct 345 PLGLQQRDVIGI 356
> cel:F01F1.7 ddx-23; DEAD boX helicase homolog family member
(ddx-23); K12858 ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=730
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 44/71 (61%), Positives = 55/71 (77%), Gaps = 0/71 (0%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
EM++RDWRIFREDF I +KGGRVP P+R W EA P E+ +A+K+ Y PTPIQ QAIP
Sbjct 274 EMSDRDWRIFREDFNISIKGGRVPRPLRNWEEAGFPDEVYQAVKEIGYLEPTPIQRQAIP 333
Query 62 IALEMRDLIGM 72
I L+ RD+IG+
Sbjct 334 IGLQNRDVIGV 344
> hsa:9416 DDX23, MGC8416, PRPF28, U5-100K, U5-100KD, prp28; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 23 (EC:3.6.4.13); K12858
ATP-dependent RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=820
Score = 99.8 bits (247), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 41/72 (56%), Positives = 56/72 (77%), Gaps = 0/72 (0%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
+EMT+RDWRIFRED+ I KGG++P PIR+W ++ LP ++E I + Y+ PTPIQ QAI
Sbjct 363 DEMTDRDWRIFREDYSITTKGGKIPNPIRSWKDSSLPPHILEVIDKCGYKEPTPIQRQAI 422
Query 61 PIALEMRDLIGM 72
PI L+ RD+IG+
Sbjct 423 PIGLQNRDIIGV 434
> mmu:74351 Ddx23, 3110082M05Rik, 4921506D17Rik; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 23 (EC:3.6.1.-); K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=819
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 41/72 (56%), Positives = 56/72 (77%), Gaps = 0/72 (0%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
+EMT+RDWRIFRED+ I KGG++P PIR+W ++ LP ++E I + Y+ PTPIQ QAI
Sbjct 362 DEMTDRDWRIFREDYSITTKGGKIPNPIRSWKDSSLPPHILEVIDKCGYKEPTPIQRQAI 421
Query 61 PIALEMRDLIGM 72
PI L+ RD+IG+
Sbjct 422 PIGLQNRDIIGV 433
> dre:334283 ddx23, wu:fi39b12, zgc:63742; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 23 (EC:3.6.1.-); K12858 ATP-dependent RNA
helicase DDX23/PRP28 [EC:3.6.4.13]
Length=807
Score = 98.2 bits (243), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 42/72 (58%), Positives = 55/72 (76%), Gaps = 0/72 (0%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
+EMT+RDWRIFRED+ I KGG++P PIR W E LP ++E I++ Y+ PTPIQ QAI
Sbjct 350 DEMTDRDWRIFREDYSITTKGGKIPNPIRNWKEYSLPPHILEVIEKCGYKDPTPIQRQAI 409
Query 61 PIALEMRDLIGM 72
PI L+ RD+IG+
Sbjct 410 PIGLQNRDIIGV 421
> cpv:cgd3_3690 U5 snRNP 100 kD protein ; K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=529
Score = 90.1 bits (222), Expect = 2e-18, Method: Composition-based stats.
Identities = 38/71 (53%), Positives = 51/71 (71%), Gaps = 0/71 (0%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
+MTERDW+IFRED+ I ++G VP PIR W + + E I+ YE+PTPIQMQ IP
Sbjct 115 DMTERDWKIFREDYSINVRGKDVPNPIRNWKDCHVLEIQTELIRNIGYEKPTPIQMQCIP 174
Query 62 IALEMRDLIGM 72
I L++RD+IG+
Sbjct 175 IGLKLRDMIGI 185
> hsa:9879 DDX46, FLJ25329, KIAA0801, MGC9936, PRPF5, Prp5; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 46 (EC:3.6.4.13); K12811
ATP-dependent RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1031
Score = 67.0 bits (162), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 49/74 (66%), Gaps = 1/74 (1%)
Query 2 EMTERDWRIFREDFE-IYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
+M++ + +FR + E I +KG P PI++W + + +++ ++K+ YE+PTPIQ QAI
Sbjct 344 KMSQEEVNVFRLEMEGITVKGKGCPKPIKSWVQCGISMKILNSLKKHGYEKPTPIQTQAI 403
Query 61 PIALEMRDLIGMQR 74
P + RDLIG+ +
Sbjct 404 PAIMSGRDLIGIAK 417
> mmu:212880 Ddx46, 2200005K02Rik, 8430438J23Rik, AI325430, AI957095,
MGC116676, MGC31579, mKIAA0801; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 46 (EC:3.6.4.13); K12811 ATP-dependent RNA
helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1031
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 28/74 (37%), Positives = 49/74 (66%), Gaps = 1/74 (1%)
Query 2 EMTERDWRIFREDFE-IYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
+M++ + +FR + E I +KG P PI++W + + +++ ++K+ YE+PTPIQ QAI
Sbjct 344 KMSQEEVNVFRLEMEGITVKGKGCPKPIKSWVQCGISMKILNSLKKHGYEKPTPIQTQAI 403
Query 61 PIALEMRDLIGMQR 74
P + RDLIG+ +
Sbjct 404 PAIMSGRDLIGIAK 417
> dre:321948 ddx46, fb39a03, wu:fb39a03; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 46 (EC:3.6.4.13); K12811 ATP-dependent RNA
helicase DDX46/PRP5 [EC:3.6.4.13]
Length=1018
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 27/58 (46%), Positives = 40/58 (68%), Gaps = 0/58 (0%)
Query 17 IYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLIGMQR 74
I +KG P PI+TW + + +++ A+K+ NYE+PTPIQ QAIP + RDLIG+ +
Sbjct 328 ISVKGKGCPKPIKTWVQCGISMKVLNALKKHNYEKPTPIQAQAIPAIMSGRDLIGIAK 385
> ath:AT1G20920 DEAD box RNA helicase, putative; K12811 ATP-dependent
RNA helicase DDX46/PRP5 [EC:3.6.4.13]
Length=828
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 47/72 (65%), Gaps = 0/72 (0%)
Query 3 MTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPI 62
MT+ + +R++ E+ + G VP PI+ W + L ++++ +K+ NYE+P PIQ QA+PI
Sbjct 165 MTQEEVNTYRKELELKVHGKDVPRPIKFWHQTGLTSKILDTMKKLNYEKPMPIQTQALPI 224
Query 63 ALEMRDLIGMQR 74
+ RD IG+ +
Sbjct 225 IMSGRDCIGVAK 236
> ath:AT5G63120 ethylene-responsive DEAD box RNA helicase, putative
(RH30)
Length=484
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 32/86 (37%), Positives = 54/86 (62%), Gaps = 2/86 (2%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
+ MTE+D ++R + +I ++G VP P++ + +A P ++EAI + + PTPIQ Q
Sbjct 137 QAMTEQDVAMYRTERDISVEGRDVPKPMKMFQDANFPDNILEAIAKLGFTEPTPIQAQGW 196
Query 61 PIALEMRDLIGMQRRDLARQQHLSYL 86
P+AL+ RDLIG+ + + L+YL
Sbjct 197 PMALKGRDLIGIA--ETGSGKTLAYL 220
> sce:YDR243C PRP28; Prp28p (EC:3.6.1.-); K12858 ATP-dependent
RNA helicase DDX23/PRP28 [EC:3.6.4.13]
Length=588
Score = 61.2 bits (147), Expect = 7e-10, Method: Composition-based stats.
Identities = 34/76 (44%), Positives = 43/76 (56%), Gaps = 5/76 (6%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAE-LPWELIEAIKQA-NYERPTPIQMQA 59
EM ERDWRI +ED+ I KGG V P+R W E +P +L+ I Q + PTPIQ
Sbjct 145 EMNERDWRILKEDYAIVTKGGTVENPLRNWEELNIIPRDLLRVIIQELRFPSPTPIQRIT 204
Query 60 IPIALEM---RDLIGM 72
IP M RD +G+
Sbjct 205 IPNVCNMKQYRDFLGV 220
> ath:AT3G09620 DEAD/DEAH box helicase, putative
Length=989
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 46/72 (63%), Gaps = 0/72 (0%)
Query 3 MTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPI 62
MT+ +R++ E+ + G VP PI+ W + L ++++ +K+ NYE+P PIQ QA+PI
Sbjct 370 MTQDAVNAYRKELELKVHGKDVPRPIQFWHQTGLTSKILDTLKKLNYEKPMPIQAQALPI 429
Query 63 ALEMRDLIGMQR 74
+ RD IG+ +
Sbjct 430 IMSGRDCIGVAK 441
> hsa:51428 DDX41, ABS, MGC8828; DEAD (Asp-Glu-Ala-Asp) box polypeptide
41 (EC:3.6.4.13); K13116 ATP-dependent RNA helicase
DDX41 [EC:3.6.4.13]
Length=622
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 42/71 (59%), Gaps = 0/71 (0%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
M+E R+ + I ++G +PPPI+++ E + P ++ +K+ PTPIQ+Q IP
Sbjct 154 SMSEERHERVRKKYHILVEGDGIPPPIKSFKEMKFPAAILRGLKKKGIHHPTPIQIQGIP 213
Query 62 IALEMRDLIGM 72
L RD+IG+
Sbjct 214 TILSGRDMIGI 224
> xla:399382 ddx5, MGC81559; DEAD (Asp-Glu-Ala-Asp) box polypeptide
5; K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=608
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 30/76 (39%), Positives = 43/76 (56%), Gaps = 2/76 (2%)
Query 11 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLI 70
+R EI ++G P PI + EA P ++EAIK+ N+ PTPIQ Q P+AL D++
Sbjct 74 YRRSKEITVRGINCPKPILNFNEASFPANVMEAIKRQNFTEPTPIQGQGWPVALSGLDMV 133
Query 71 GMQRRDLARQQHLSYL 86
G+ + LSYL
Sbjct 134 GVAMTGSGKT--LSYL 147
> mmu:72935 Ddx41, 2900024F02Rik, AA958953, ABS, AI324246; DEAD
(Asp-Glu-Ala-Asp) box polypeptide 41 (EC:3.6.4.13); K13116
ATP-dependent RNA helicase DDX41 [EC:3.6.4.13]
Length=622
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 42/71 (59%), Gaps = 0/71 (0%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
M+E R+ + I ++G +PPPI+++ E + P ++ +K+ PTPIQ+Q IP
Sbjct 154 SMSEERHERVRKKYHILVEGDGIPPPIKSFKEMKFPAAILRGLKKKGILHPTPIQIQGIP 213
Query 62 IALEMRDLIGM 72
L RD+IG+
Sbjct 214 TILSGRDMIGI 224
> ath:AT1G55150 DEAD box RNA helicase, putative (RH20); K12823
ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=501
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 52/84 (61%), Gaps = 2/84 (2%)
Query 3 MTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPI 62
MT+ + +R+ EI ++G +P P++++ + P ++E +K+A + PTPIQ Q P+
Sbjct 73 MTDTEVEEYRKLREITVEGKDIPKPVKSFRDVGFPDYVLEEVKKAGFTEPTPIQSQGWPM 132
Query 63 ALEMRDLIGMQRRDLARQQHLSYL 86
A++ RDLIG+ + + LSYL
Sbjct 133 AMKGRDLIGIA--ETGSGKTLSYL 154
> ath:AT2G42520 DEAD box RNA helicase, putative; K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=633
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 44/87 (50%), Gaps = 4/87 (4%)
Query 3 MTERDWRIFR----EDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQ 58
TE+D + ED I G VPPP+ T+AE +L L I++ Y +PTP+Q
Sbjct 129 FTEQDNTVINFDAYEDIPIETSGDNVPPPVNTFAEIDLGEALNLNIRRCKYVKPTPVQRH 188
Query 59 AIPIALEMRDLIGMQRRDLARQQHLSY 85
AIPI LE RDL+ + + +
Sbjct 189 AIPILLEGRDLMACAQTGSGKTAAFCF 215
> cel:H27M09.1 hypothetical protein; K13116 ATP-dependent RNA
helicase DDX41 [EC:3.6.4.13]
Length=630
Score = 58.5 bits (140), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 43/70 (61%), Gaps = 1/70 (1%)
Query 4 TERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAI-KQANYERPTPIQMQAIPI 62
++ D+ I R+ I +G +PPPI ++ E + P L+E + KQ PT IQ+Q IP+
Sbjct 164 SQEDYEIQRKRLGISCEGDHIPPPIGSFLEMKFPKSLLEFMQKQKGIVTPTAIQIQGIPV 223
Query 63 ALEMRDLIGM 72
AL RD+IG+
Sbjct 224 ALSGRDMIGI 233
> ath:AT4G33370 DEAD-box protein abstrakt, putative
Length=542
Score = 57.4 bits (137), Expect = 1e-08, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 42/71 (59%), Gaps = 0/71 (0%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
+M+ + + R+ + I + G +PPPI+ + + + P L+ +K PTPIQ+Q +P
Sbjct 70 KMSTKQMDLIRKQWHITVNGEDIPPPIKNFMDMKFPSPLLRMLKDKGIMHPTPIQVQGLP 129
Query 62 IALEMRDLIGM 72
+ L RD+IG+
Sbjct 130 VVLSGRDMIGI 140
> ath:AT3G58510 DEAD box RNA helicase, putative (RH11); K11594
ATP-dependent RNA helicase [EC:3.6.4.13]
Length=612
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 39/73 (53%), Gaps = 0/73 (0%)
Query 13 EDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLIGM 72
ED + GG VPPP+ T+A+ +L L I++ Y RPTP+Q AIPI L RDL+
Sbjct 135 EDIPVETSGGDVPPPVNTFADIDLGDALNLNIRRCKYVRPTPVQRHAIPILLAERDLMAC 194
Query 73 QRRDLARQQHLSY 85
+ + +
Sbjct 195 AQTGSGKTAAFCF 207
> ath:AT5G51280 DEAD-box protein abstrakt, putative; K13116 ATP-dependent
RNA helicase DDX41 [EC:3.6.4.13]
Length=591
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 45/71 (63%), Gaps = 0/71 (0%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
+M+ + + R+ + I + G +PPPI+ + + + P +++ +K+ +PTPIQ+Q +P
Sbjct 119 KMSSKQRDLIRKQWHIIVNGDDIPPPIKNFKDMKFPRPVLDTLKEKGIVQPTPIQVQGLP 178
Query 62 IALEMRDLIGM 72
+ L RD+IG+
Sbjct 179 VILAGRDMIGI 189
> dre:556764 similar to Probable RNA-dependent helicase p72 (DEAD-box
protein p72) (DEAD-box protein 17); K13178 ATP-dependent
RNA helicase DDX17 [EC:3.6.4.13]
Length=671
Score = 57.4 bits (137), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 48/86 (55%), Gaps = 2/86 (2%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
M++ D +R EI ++G P P+ + +A+ P +++ + Q N++ PT IQ Q
Sbjct 67 HHMSQYDVEEYRRKREITVRGSGCPKPVTNFHQAQFPQYVMDVLLQQNFKEPTAIQAQGF 126
Query 61 PIALEMRDLIGMQRRDLARQQHLSYL 86
P+AL RD++G+ + + L+YL
Sbjct 127 PLALSGRDMVGIAQTGSGKT--LAYL 150
> xla:379390 MGC53795; similar to DEAD/H (Asp-Glu-Ala-Asp/His)
box polypeptide 5 (RNA helicase, 68kDa); K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=607
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 42/76 (55%), Gaps = 2/76 (2%)
Query 11 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLI 70
+R EI ++G P P+ + EA P ++E IK+ N+ PTPIQ Q P+AL D++
Sbjct 72 YRRSKEITVRGLNCPKPVLNFHEASFPANVMEVIKRLNFTEPTPIQGQGWPVALSGLDMV 131
Query 71 GMQRRDLARQQHLSYL 86
G+ + LSYL
Sbjct 132 GVAMTGSGKT--LSYL 145
> ath:AT3G02065 DEAD/DEAH box helicase family protein
Length=505
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 27/70 (38%), Positives = 43/70 (61%), Gaps = 2/70 (2%)
Query 4 TERDWRIFREDFEIYLKG--GRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
+ D ++ R +I+++G VPPP+ T+ LP +L+ ++ A Y+ PTPIQMQAIP
Sbjct 83 SSHDAQLLRRKLDIHVQGQGSAVPPPVLTFTSCGLPPKLLLNLETAGYDFPTPIQMQAIP 142
Query 62 IALEMRDLIG 71
AL + L+
Sbjct 143 AALTGKSLLA 152
> ath:AT3G01540 DRH1; DRH1 (DEAD BOX RNA HELICASE 1); ATP-dependent
RNA helicase/ ATPase
Length=619
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 42/71 (59%), Gaps = 2/71 (2%)
Query 16 EIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLIGMQRR 75
EI + GG+VPPP+ ++ P EL+ + A + PTPIQ Q+ PIA++ RD++ + +
Sbjct 145 EITVSGGQVPPPLMSFEATGFPPELLREVLSAGFSAPTPIQAQSWPIAMQGRDIVAIAKT 204
Query 76 DLARQQHLSYL 86
+ L YL
Sbjct 205 GSGKT--LGYL 213
> cel:F53H1.1 hypothetical protein; K12811 ATP-dependent RNA helicase
DDX46/PRP5 [EC:3.6.4.13]
Length=970
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 27/73 (36%), Positives = 47/73 (64%), Gaps = 1/73 (1%)
Query 3 MTERDWRIFREDFE-IYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
MT+ + + +RE+ + I +KG P PI+TWA+ + +++ +K+ Y +PT IQ QAIP
Sbjct 277 MTKAEVKAYREELDSITVKGIDCPKPIKTWAQCGVNLKMMNVLKKFEYSKPTSIQAQAIP 336
Query 62 IALEMRDLIGMQR 74
+ RD+IG+ +
Sbjct 337 SIMSGRDVIGIAK 349
> ath:AT2G47330 DEAD/DEAH box helicase, putative; K12835 ATP-dependent
RNA helicase DDX42 [EC:3.6.4.13]
Length=760
Score = 56.2 bits (134), Expect = 2e-08, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 45/72 (62%), Gaps = 0/72 (0%)
Query 3 MTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPI 62
MTE++ +R+ I + G V P++T+ + +++ AIK+ YE+PT IQ QA+PI
Sbjct 202 MTEQETTDYRQRLGIRVSGFDVHRPVKTFEDCGFSSQIMSAIKKQAYEKPTAIQCQALPI 261
Query 63 ALEMRDLIGMQR 74
L RD+IG+ +
Sbjct 262 VLSGRDVIGIAK 273
> ath:AT1G28180 ATP binding / ATP-dependent helicase/ helicase/
nucleic acid binding
Length=614
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 28/72 (38%), Positives = 36/72 (50%), Gaps = 25/72 (34%)
Query 1 EEMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAI 60
EEM ERDWRIF+EDF I +G ++P P+R W E I
Sbjct 199 EEMNERDWRIFKEDFNISYRGSKIPHPMRNWEE-------------------------TI 233
Query 61 PIALEMRDLIGM 72
P+ LE RD+IG+
Sbjct 234 PLGLEQRDVIGI 245
> xla:398649 ddx17, MGC80019; DEAD (Asp-Glu-Ala-Asp) box polypeptide
17; K13178 ATP-dependent RNA helicase DDX17 [EC:3.6.4.13]
Length=610
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 45/84 (53%), Gaps = 2/84 (2%)
Query 3 MTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPI 62
MT+ D R EI ++G P P+ + +A P +++ + ++ PTPIQ Q P+
Sbjct 56 MTQHDVEELRRKKEITIRGVNCPKPLYAFHQANFPQYVLDVLLDQRFKEPTPIQCQGFPL 115
Query 63 ALEMRDLIGMQRRDLARQQHLSYL 86
AL RD++G+ + + L+YL
Sbjct 116 ALSGRDMVGIAQTGSGKT--LAYL 137
> sce:YNL112W DBP2; Dbp2p (EC:3.6.1.-); K12823 ATP-dependent RNA
helicase DDX5/DBP2 [EC:3.6.4.13]
Length=546
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 43/76 (56%), Gaps = 2/76 (2%)
Query 11 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLI 70
FR++ E+ + G +P PI T+ EA P ++ +K +++PT IQ Q P+AL RD++
Sbjct 95 FRKENEMTISGHDIPKPITTFDEAGFPDYVLNEVKAEGFDKPTGIQCQGWPMALSGRDMV 154
Query 71 GMQRRDLARQQHLSYL 86
G+ + LSY
Sbjct 155 GIAA--TGSGKTLSYC 168
> bbo:BBOV_II004470 18.m06373; p68-like protein; K12823 ATP-dependent
RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=529
Score = 54.7 bits (130), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 28/71 (39%), Positives = 43/71 (60%), Gaps = 1/71 (1%)
Query 3 MTERDWRIFREDFEIYLKGGR-VPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
M+ D R++ EI + GR VP P+ ++ P +++AI+ A + PTPIQ+Q P
Sbjct 81 MSSADVDRVRKEREITIIAGRDVPKPVVSFEHTSFPDYILKAIRAAGFTAPTPIQVQGWP 140
Query 62 IALEMRDLIGM 72
IAL RD+IG+
Sbjct 141 IALSGRDVIGI 151
> dre:394020 ddx41, MGC55896, wu:fb92e02, zgc:55896; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 41 (EC:3.6.4.13); K13116 ATP-dependent
RNA helicase DDX41 [EC:3.6.4.13]
Length=306
Score = 54.3 bits (129), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 39/61 (63%), Gaps = 0/61 (0%)
Query 12 REDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLIG 71
R+ + I ++G +P PI+++ E + P +++ +K+ PTPIQ+Q IP L RD+IG
Sbjct 155 RKKYHILVEGEGIPAPIKSFREMKFPQAILKGLKKKGIVHPTPIQIQGIPTILSGRDMIG 214
Query 72 M 72
+
Sbjct 215 I 215
> dre:322206 ddx5, wu:fa56a07, wu:fb11e01, wu:fb16c10, wu:fb53b05;
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.1.-);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=518
Score = 54.3 bits (129), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 28/76 (36%), Positives = 43/76 (56%), Gaps = 2/76 (2%)
Query 11 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLI 70
+R EI +KG P PI + EA P +++ I + N+ PTPIQ Q P+AL +D++
Sbjct 78 YRRSKEITVKGRDGPKPIVKFHEANFPKYVMDVITKQNWTDPTPIQAQGWPVALSGKDMV 137
Query 71 GMQRRDLARQQHLSYL 86
G+ + + LSYL
Sbjct 138 GIAQTGSGKT--LSYL 151
> hsa:1655 DDX5, DKFZp434E109, DKFZp686J01190, G17P1, HLR1, HUMP68,
p68; DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.4.13);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=614
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 44/83 (53%), Gaps = 2/83 (2%)
Query 4 TERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIA 63
T ++ +R EI ++G P P+ + EA P +++ I + N+ PT IQ Q P+A
Sbjct 69 TAQEVETYRRSKEITVRGHNCPKPVLNFYEANFPANVMDVIARQNFTEPTAIQAQGWPVA 128
Query 64 LEMRDLIGMQRRDLARQQHLSYL 86
L D++G+ + + LSYL
Sbjct 129 LSGLDMVGVAQTGSGKT--LSYL 149
> mmu:13207 Ddx5, 2600009A06Rik, G17P1, HUMP68, Hlr1, MGC118083,
p68; DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (EC:3.6.4.13);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=615
Score = 53.5 bits (127), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 44/83 (53%), Gaps = 2/83 (2%)
Query 4 TERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIA 63
T ++ +R EI ++G P P+ + EA P +++ I + N+ PT IQ Q P+A
Sbjct 69 TAQEVDTYRRSKEITVRGHNCPKPVLNFYEANFPANVMDVIARQNFTEPTAIQAQGWPVA 128
Query 64 LEMRDLIGMQRRDLARQQHLSYL 86
L D++G+ + + LSYL
Sbjct 129 LSGLDMVGVAQTGSGKT--LSYL 149
> tpv:TP02_0316 RNA helicase-1; K13116 ATP-dependent RNA helicase
DDX41 [EC:3.6.4.13]
Length=598
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 38/61 (62%), Gaps = 0/61 (0%)
Query 12 REDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLIG 71
R I + G +VPPPI T+ + +LP +++A++ PT IQMQA+P L RD+IG
Sbjct 175 RNALVIDVSGDQVPPPILTFEDMKLPRPILKALRHKKIFEPTKIQMQAMPAVLLGRDVIG 234
Query 72 M 72
+
Sbjct 235 I 235
> xla:446276 ddx52; DEAD (Asp-Glu-Ala-Asp) box polypeptide 52;
K14779 ATP-dependent RNA helicase DDX52/ROK1 [EC:3.6.4.13]
Length=614
Score = 53.5 bits (127), Expect = 2e-07, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 40/65 (61%), Gaps = 4/65 (6%)
Query 11 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAI----KQANYERPTPIQMQAIPIALEM 66
FR + +IY++G +P P T+ + E ++++ I K A + PTPIQMQAIPI L
Sbjct 149 FRNEHKIYVQGTDIPEPAATFQQLEQEYKILSKIMQNVKDAGFHTPTPIQMQAIPIMLHD 208
Query 67 RDLIG 71
R+++
Sbjct 209 REILA 213
> cpv:cgd7_4600 abstrakt protein SF II helicase + Znknuckle C2HC
(PA) ; K13116 ATP-dependent RNA helicase DDX41 [EC:3.6.4.13]
Length=570
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 24/62 (38%), Positives = 36/62 (58%), Gaps = 0/62 (0%)
Query 11 FREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLI 70
R I + G VPPPI ++ + P E+++A+ +P+ IQMQ +PI L RDLI
Sbjct 90 LRSRLLIVVNGSDVPPPILSFKDMGFPQEILDALASKGISKPSQIQMQGLPIILMGRDLI 149
Query 71 GM 72
G+
Sbjct 150 GL 151
> mmu:432554 Gm12183, OTTMUSG00000005521; predicted gene 12183;
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=670
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 44/83 (53%), Gaps = 2/83 (2%)
Query 4 TERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIA 63
T ++ +R EI ++G P P+ + EA P +++ I + N+ PT IQ Q P+A
Sbjct 124 TAQEVDTYRRSKEITVRGHNCPKPVLKFYEANFPANVMDVIARQNFTEPTAIQAQGWPVA 183
Query 64 LEMRDLIGMQRRDLARQQHLSYL 86
L D++G+ + + LSYL
Sbjct 184 LSGLDMVGVAQTGSGKT--LSYL 204
> xla:379217 MGC53409; similar to ATP-dependent, RNA helicase;
K14779 ATP-dependent RNA helicase DDX52/ROK1 [EC:3.6.4.13]
Length=686
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 41/65 (63%), Gaps = 4/65 (6%)
Query 11 FREDFEIYLKGGRVPPPIRTWAEAELPWEL----IEAIKQANYERPTPIQMQAIPIALEM 66
FR + +IY++G +P P T+ + E +++ ++ +K A + PTPIQMQAIPI L
Sbjct 149 FRNEQKIYIQGTDIPEPAATFQQLEQEYKIHSKIMQNVKDAGFHTPTPIQMQAIPIMLHG 208
Query 67 RDLIG 71
R+++
Sbjct 209 REILA 213
> ath:AT3G06480 DEAD box RNA helicase, putative
Length=1088
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 42/77 (54%), Gaps = 2/77 (2%)
Query 10 IFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDL 69
I+R+ E+ G +P P T+ + LP E++ + A + PTPIQ Q PIAL+ RD+
Sbjct 416 IYRKQHEVTTTGENIPAPYITFESSGLPPEILRELLSAGFPSPTPIQAQTWPIALQSRDI 475
Query 70 IGMQRRDLARQQHLSYL 86
+ + + + L YL
Sbjct 476 VAIAKTGSGKT--LGYL 490
> ath:AT3G58570 DEAD box RNA helicase, putative; K11594 ATP-dependent
RNA helicase [EC:3.6.4.13]
Length=646
Score = 51.6 bits (122), Expect = 6e-07, Method: Composition-based stats.
Identities = 26/59 (44%), Positives = 34/59 (57%), Gaps = 0/59 (0%)
Query 13 EDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRDLIG 71
ED I G VPPP+ T+AE +L L I++ Y +PTP+Q AIPI RDL+
Sbjct 130 EDIPIETSGDNVPPPVNTFAEIDLGEALNLNIQRCKYVKPTPVQRNAIPILAAGRDLMA 188
> tgo:TGME49_010360 DEAD/DEAH box helicase, putative (EC:5.99.1.3);
K13116 ATP-dependent RNA helicase DDX41 [EC:3.6.4.13]
Length=657
Score = 51.2 bits (121), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 39/71 (54%), Gaps = 0/71 (0%)
Query 2 EMTERDWRIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIP 61
EMT + RE F I + G PPP R + + P +++ +++ PT IQMQ IP
Sbjct 181 EMTLAEANEVRERFFIDVSGEDPPPPFRNFKDMRFPQPILKGLQERGISYPTQIQMQGIP 240
Query 62 IALEMRDLIGM 72
L+ RD+IG+
Sbjct 241 AILQGRDIIGI 251
> tgo:TGME49_036650 DEAD/DEAH box helicase, putative (EC:5.99.1.3);
K12823 ATP-dependent RNA helicase DDX5/DBP2 [EC:3.6.4.13]
Length=550
Score = 50.8 bits (120), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 37/64 (57%), Gaps = 0/64 (0%)
Query 9 RIFREDFEIYLKGGRVPPPIRTWAEAELPWELIEAIKQANYERPTPIQMQAIPIALEMRD 68
RI R + + G VP P+ T+ P +++ I Q +++PT IQ+Q PIAL RD
Sbjct 109 RIRRANEITIVHGHNVPKPVPTFEYTSFPSYILDVINQTGFQKPTAIQVQGWPIALSGRD 168
Query 69 LIGM 72
+IG+
Sbjct 169 MIGI 172
> mmu:100048658 Ddx43, OTTMUSG00000019690; DEAD (Asp-Glu-Ala-Asp)
box polypeptide 43 (EC:3.6.4.13)
Length=646
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 11/87 (12%)
Query 7 DWRIFREDFEIY---LKGGR---VPPPIRTWAEAELPW-ELIEAIKQANYERPTPIQMQA 59
+WR +E+F I LK G +P PI + +A + E++E IK+A +++PTPIQ QA
Sbjct 213 NWR--KENFNITCDDLKDGEKRPIPNPICKFEDAFQSYPEVMENIKRAGFQKPTPIQSQA 270
Query 60 IPIALEMRDLIGMQRRDLARQQHLSYL 86
PI L+ DLIG+ + + LSYL
Sbjct 271 WPIVLQGIDLIGVAQTGTGKT--LSYL 295
Lambda K H
0.322 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2026251472
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40