bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
164,496 sequences; 82,071,388 total letters
Query= Emax_3374_orf3
Length=68
Score E
Sequences producing significant alignments: (Bits) Value
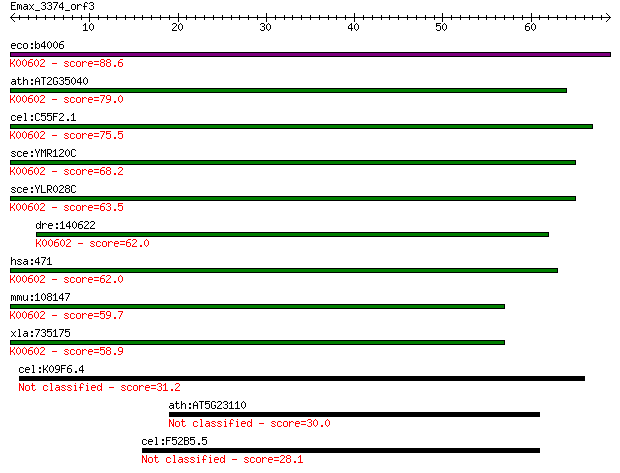
eco:b4006 purH, ECK3998, JW3970; fused IMP cyclohydrolase/phos... 88.6 5e-18
ath:AT2G35040 AICARFT/IMPCHase bienzyme family protein (EC:3.5... 79.0 4e-15
cel:C55F2.1 hypothetical protein; K00602 phosphoribosylaminoim... 75.5 4e-14
sce:YMR120C ADE17; Ade17p (EC:3.5.4.10 2.1.2.3); K00602 phosph... 68.2 7e-12
sce:YLR028C ADE16; Ade16p (EC:3.5.4.10 2.1.2.3); K00602 phosph... 63.5 2e-10
dre:140622 atic, cb72, id:ibd1333, wu:fb50b10, wu:fb58a11, wu:... 62.0 4e-10
hsa:471 ATIC, AICAR, AICARFT, FLJ93545, IMPCHASE, PURH; 5-amin... 62.0 4e-10
mmu:108147 Atic, 2610509C24Rik, AA536954, AW212393; 5-aminoimi... 59.7 2e-09
xla:735175 atic, MGC130953; 5-aminoimidazole-4-carboxamide rib... 58.9 4e-09
cel:K09F6.4 hypothetical protein 31.2 0.91
ath:AT5G23110 zinc finger (C3HC4-type RING finger) family protein 30.0 1.8
cel:F52B5.5 cep-1; P-53-like protein family member (cep-1) 28.1 6.8
> eco:b4006 purH, ECK3998, JW3970; fused IMP cyclohydrolase/phosphoribosylaminoimidazolecarboxamide
formyltransferase (EC:3.5.4.10
2.1.2.3); K00602 phosphoribosylaminoimidazolecarboxamide
formyltransferase / IMP cyclohydrolase [EC:2.1.2.3 3.5.4.10]
Length=529
Score = 88.6 bits (218), Expect = 5e-18, Method: Composition-based stats.
Identities = 44/70 (62%), Positives = 52/70 (74%), Gaps = 2/70 (2%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V VNLYPF T+ + GC L A+ENIDIGGPTMVRSAAKNHKDVAIVV +SDY +++ +
Sbjct 101 VVVNLYPFAQTVAREGCSLEDAVENIDIGGPTMVRSAAKNHKDVAIVVKSSDYDAIIKEM 160
Query 61 --KAGGLTYA 68
G LT A
Sbjct 161 DDNEGSLTLA 170
> ath:AT2G35040 AICARFT/IMPCHase bienzyme family protein (EC:3.5.4.10);
K00602 phosphoribosylaminoimidazolecarboxamide formyltransferase
/ IMP cyclohydrolase [EC:2.1.2.3 3.5.4.10]
Length=596
Score = 79.0 bits (193), Expect = 4e-15, Method: Composition-based stats.
Identities = 39/64 (60%), Positives = 44/64 (68%), Gaps = 1/64 (1%)
Query 1 VAVNLYPFEATINKPG-CDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLES 59
V VNLYPF + PG IENIDIGGP M+R+AAKNHKDV IVV++ DY VLE
Sbjct 167 VVVNLYPFYEKVTAPGGISFEDGIENIDIGGPAMIRAAAKNHKDVLIVVDSGDYQAVLEY 226
Query 60 LKAG 63
LK G
Sbjct 227 LKGG 230
> cel:C55F2.1 hypothetical protein; K00602 phosphoribosylaminoimidazolecarboxamide
formyltransferase / IMP cyclohydrolase
[EC:2.1.2.3 3.5.4.10]
Length=594
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 48/66 (72%), Gaps = 0/66 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V NLYPF+ T+ C + A+ENIDIGG T++R+AAKNH+ V+++ + +DY +++ L
Sbjct 100 VVCNLYPFKKTVQSKDCSVEEAVENIDIGGVTLLRAAAKNHERVSVICDPADYDHIISEL 159
Query 61 KAGGLT 66
K+GG T
Sbjct 160 KSGGTT 165
> sce:YMR120C ADE17; Ade17p (EC:3.5.4.10 2.1.2.3); K00602 phosphoribosylaminoimidazolecarboxamide
formyltransferase / IMP
cyclohydrolase [EC:2.1.2.3 3.5.4.10]
Length=592
Score = 68.2 bits (165), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 31/64 (48%), Positives = 42/64 (65%), Gaps = 0/64 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V NLYPF+ T+ K G +P A+E IDIGG T++R+AAKNH V I+ + DY+ L L
Sbjct 100 VVCNLYPFKETVAKVGVTIPEAVEEIDIGGVTLLRAAAKNHARVTILSDPKDYSEFLSEL 159
Query 61 KAGG 64
+ G
Sbjct 160 SSNG 163
> sce:YLR028C ADE16; Ade16p (EC:3.5.4.10 2.1.2.3); K00602 phosphoribosylaminoimidazolecarboxamide
formyltransferase / IMP
cyclohydrolase [EC:2.1.2.3 3.5.4.10]
Length=591
Score = 63.5 bits (153), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 30/64 (46%), Positives = 42/64 (65%), Gaps = 0/64 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
V NLYPF+ T+ K G + A+E IDIGG T++R+AAKNH V I+ + +DY+ L+ L
Sbjct 100 VVCNLYPFKETVAKIGVTVQEAVEEIDIGGVTLLRAAAKNHSRVTILSDPNDYSIFLQDL 159
Query 61 KAGG 64
G
Sbjct 160 SKDG 163
> dre:140622 atic, cb72, id:ibd1333, wu:fb50b10, wu:fb58a11, wu:fi76g09,
wu:fy77g01; 5-aminoimidazole-4-carboxamide ribonucleotide
formyltransferase/IMP cyclohydrolase; K00602 phosphoribosylaminoimidazolecarboxamide
formyltransferase / IMP cyclohydrolase
[EC:2.1.2.3 3.5.4.10]
Length=590
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 28/58 (48%), Positives = 40/58 (68%), Gaps = 0/58 (0%)
Query 4 NLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESLK 61
NLYPF T++ PG + A+E IDIGG T++R+AAKNH V +V + SDY V + ++
Sbjct 101 NLYPFVKTVSSPGVTVEDAVEQIDIGGVTLLRAAAKNHARVTVVCDPSDYNVVAKEME 158
> hsa:471 ATIC, AICAR, AICARFT, FLJ93545, IMPCHASE, PURH; 5-aminoimidazole-4-carboxamide
ribonucleotide formyltransferase/IMP
cyclohydrolase (EC:2.1.2.3 3.5.4.10); K00602 phosphoribosylaminoimidazolecarboxamide
formyltransferase / IMP cyclohydrolase
[EC:2.1.2.3 3.5.4.10]
Length=592
Score = 62.0 bits (149), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/62 (46%), Positives = 39/62 (62%), Gaps = 0/62 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
VA NLYPF T+ PG + A+E IDIGG T++R+AAKNH V +V DY V +
Sbjct 99 VACNLYPFVKTVASPGVTVEEAVEQIDIGGVTLLRAAAKNHARVTVVCEPEDYVVVSTEM 158
Query 61 KA 62
++
Sbjct 159 QS 160
> mmu:108147 Atic, 2610509C24Rik, AA536954, AW212393; 5-aminoimidazole-4-carboxamide
ribonucleotide formyltransferase/IMP
cyclohydrolase (EC:2.1.2.3 3.5.4.10); K00602 phosphoribosylaminoimidazolecarboxamide
formyltransferase / IMP cyclohydrolase
[EC:2.1.2.3 3.5.4.10]
Length=592
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/56 (50%), Positives = 35/56 (62%), Gaps = 0/56 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANV 56
V NLYPF T+ P + A+E IDIGG T++R+AAKNH V +V DYA V
Sbjct 99 VVCNLYPFVKTVASPDVTVEAAVEQIDIGGVTLLRAAAKNHARVTVVCEPEDYAGV 154
> xla:735175 atic, MGC130953; 5-aminoimidazole-4-carboxamide ribonucleotide
formyltransferase/IMP cyclohydrolase; K00602 phosphoribosylaminoimidazolecarboxamide
formyltransferase / IMP
cyclohydrolase [EC:2.1.2.3 3.5.4.10]
Length=589
Score = 58.9 bits (141), Expect = 4e-09, Method: Composition-based stats.
Identities = 26/56 (46%), Positives = 38/56 (67%), Gaps = 0/56 (0%)
Query 1 VAVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANV 56
V NLYPF T++ G + A+E IDIGG T++R+AAKNH V ++ + SDY ++
Sbjct 97 VVCNLYPFVKTVSAAGVTVEDAVEQIDIGGVTLLRAAAKNHARVTVLCDPSDYKSI 152
> cel:K09F6.4 hypothetical protein
Length=709
Score = 31.2 bits (69), Expect = 0.91, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 10/65 (15%)
Query 2 AVNLYPFEATINKPGCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLE-SL 60
A+N+ F A + K +L NI +GGP +R + + +DYA++ + +L
Sbjct 474 AINMVEFGARLKKLKTELEIWKSNITVGGPHRIRRS---------TTSLADYAHIFQNAL 524
Query 61 KAGGL 65
K GG+
Sbjct 525 KVGGV 529
> ath:AT5G23110 zinc finger (C3HC4-type RING finger) family protein
Length=4706
Score = 30.0 bits (66), Expect = 1.8, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Query 19 LPTAIENI--DIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
LP A+ N+ IGG + + H D++ V+ + Y VLES+
Sbjct 725 LPAAVRNVLEKIGGKILNNNIKVEHSDLSSFVSDASYTGVLESI 768
> cel:F52B5.5 cep-1; P-53-like protein family member (cep-1)
Length=644
Score = 28.1 bits (61), Expect = 6.8, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 16 GCDLPTAIENIDIGGPTMVRSAAKNHKDVAIVVNASDYANVLESL 60
GC P E+ + P+M RS N+ + ++ ++Y V+E L
Sbjct 505 GCVPPIETEHENCQSPSMKRSRCTNYSFRTLTLSTAEYTKVVEFL 549
Lambda K H
0.314 0.132 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2033830404
Database: egene_temp_file_orthology_annotation_similarity_blast_database_865
Posted date: Sep 17, 2011 11:19 AM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40