bitscore colors: <40, 40-50 , 50-80, 80-200, >200
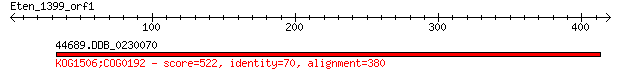
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_1399_orf1
Length=419
Score E
Sequences producing significant alignments: (Bits) Value
44689.DDB_0230070 522 1e-146
> 44689.DDB_0230070
Length=384
Score = 522 bits (1344), Expect = 1e-146, Method: Compositional matrix adjust.
Identities = 266/381 (69%), Positives = 308/381 (80%), Gaps = 3/381 (0%)
Query 33 FLFTSESVNEGHPDKLCDQVSDAILDACLAEDPNSKVACETCTKTGMVMIFGEITTNANV 92
+LFTSESV EGHPDK+CDQVSDA+LDACLA+DP SKVACET TKTGMVMI GEITT ANV
Sbjct 4 YLFTSESVTEGHPDKICDQVSDAVLDACLAQDPLSKVACETATKTGMVMILGEITTKANV 63
Query 93 DYEKIIRETCRDIGYDDESKGLDYKTMKVVVAIEDQSPEIAQAVYVNK-NIEEIGAGDQG 151
DY+K++RE + IGYDD SKG DYKT V+VAIE QSP+IAQ V++ K N E+IGAGDQG
Sbjct 64 DYQKVVREAVKKIGYDDSSKGFDYKTCNVLVAIEQQSPDIAQGVHIGKVNPEDIGAGDQG 123
Query 152 HMFGYACDETPELMPLSHSLATNLGKRLTDVRKSGVLPYIRPDGKTQVTVEYDNTSGVPV 211
HMFGYA +ETP LMPL+H LA+ L +LT++R +G L + RPD KTQVTVEY
Sbjct 124 HMFGYATNETPTLMPLTHFLASELVNKLTELRHNGTLAWARPDAKTQVTVEYKKEGHKIT 183
Query 212 PVRVHTIVISTQHAPEVANEQLRRDIMEHVIKPVVPAKYLDDNTIYHLNPSGRFVIGGPH 271
PVRVHTIVISTQH +V NEQ+R D+ E VIK VVPA+YLDDNTIYHLNPSGRFVIGGP
Sbjct 184 PVRVHTIVISTQHDEKVTNEQIRADLKELVIKAVVPAQYLDDNTIYHLNPSGRFVIGGPM 243
Query 272 GDAGLTGRKIIIDTYGGWGAHGGGAFSGKDSTKVDRSAAYAARWAAKSLVANGLCKRCLV 331
GD+GLTGRKIIID+YGGWGAHGGGAFSGKD+TKVDRS AYAARW AKSLVA GL RCLV
Sbjct 244 GDSGLTGRKIIIDSYGGWGAHGGGAFSGKDATKVDRSGAYAARWIAKSLVAAGLADRCLV 303
Query 332 QVSYSIGVAAPLSLFVDSYGTCAEGYTDDCLLRIVAENFDFRPGVIQRDLRLKEPLFKEL 391
QVSYSIGVA PLS+FVD+YGT + TD +L I+ NFD RPG + RDL+L P+F++
Sbjct 304 QVSYSIGVAKPLSVFVDTYGTGKK--TDAEILAIIDNNFDLRPGCLMRDLQLTRPIFQKT 361
Query 392 AAYGHFGRCPEKYAWEKPRDL 412
A+ GHFGR + WE ++L
Sbjct 362 ASGGHFGRNDPDFTWETVKEL 382
Lambda K H
0.320 0.137 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 169961605920
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40