bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_1791_orf1
Length=590
Score E
Sequences producing significant alignments: (Bits) Value
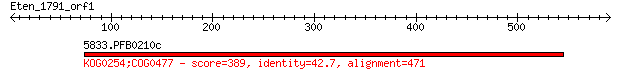
5833.PFB0210c 389 2e-106
> 5833.PFB0210c
Length=504
Score = 389 bits (998), Expect = 2e-106, Method: Compositional matrix adjust.
Identities = 201/471 (42%), Positives = 291/471 (61%), Gaps = 3/471 (0%)
Query 74 QFVLVAVLGSFQFGYSFSALNTSKAIIIADFDWCKGDANHFIDCSNGVLYGSLITTAVFI 133
++VL A + SF FGY S LNT K I+ +F+WCKG+ + ++CSN + S + +VFI
Sbjct 28 KYVLSACIASFIFGYQVSVLNTIKNFIVVEFEWCKGEKDR-LNCSNNTIQSSFLLASVFI 86
Query 134 GLTFGCLLAGPVTNFGRRVALILTNLLFLVSSALAAAAEGVASLFLARLYQGLAVGLATV 193
G GC +G + FGRR++L++ F + S L + ++ ARL G +GL TV
Sbjct 87 GAVLGCGFSGYLVQFGRRLSLLIIYNFFFLVSILTSITHHFHTILFARLLSGFGIGLVTV 146
Query 194 CVPMYISEFTPDSSRGFYGTLHQLLITIGILVATLLGLAFGSPPTDTDFTYKVSLFQQCW 253
VPMYISE T +G YG +HQL IT GI VA +LGLA G P D T ++ F + W
Sbjct 147 SVPMYISEMTHKDKKGAYGVMHQLFITFGIFVAVMLGLAMGEGP-KADSTEPLTSFAKLW 205
Query 254 WRFMLAFPAAVSIVAILLLWLVYTTESPHFLMHQGRKNTATALLREILGKEDVSHEVQTI 313
WR M FP+ +S++ IL L + + E+P+FL +GR + +L++I ++V + I
Sbjct 206 WRLMFLFPSVISLIGILALVVFFKEETPYFLFEKGRIEESKNILKKIYETDNVDEPLNAI 265
Query 314 DVAICQQKVLEAESLSLGKAMTFPIYRHVLLLAFFLSAFQQFTGINILVANSNKLYTSLN 373
A+ Q + + SLSL A+ P YR+V++L LS QQFTGIN+LV+NSN+LY
Sbjct 266 KEAVEQNESAKKNSLSLLSALKIPSYRYVIILGCLLSGLQQFTGINVLVSNSNELYKEF- 324
Query 374 IDASLVTGLSVVFGFVNVLMTVPAIFLMDRLGRRTLLLSGCFGQAASTIIALIANLVDRH 433
+D+ L+T LSVV VN LMT PAI+++++LGR+TLLL GC G + + IAN ++R+
Sbjct 325 LDSHLITILSVVMTAVNFLMTFPAIYIVEKLGRKTLLLWGCVGVLVAYLPTAIANEINRN 384
Query 434 NTGVQWLTVACIYVFVISFAVGYGPVLWVYLHEIFPPEIKQTGASVASAINAVATIIVVL 493
+ V+ L++ +V +ISFAV YGPVLW+YLHE+FP EIK + AS+AS +N V IIVV
Sbjct 385 SNFVKILSIVATFVMIISFAVSYGPVLWIYLHEMFPSEIKDSAASLASLVNWVCAIIVVF 444
Query 494 PSDFVLTSDMRVMLGICGGTSLIAFIISFIFMKETAGLSIDESPYFKGKRR 544
PSD ++ ++ + S++ F F F+KET G I SPY + R
Sbjct 445 PSDIIIKKSPSILFIVFSVMSILTFFFIFFFIKETKGGEIGTSPYITMEER 495
Lambda K H
0.327 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 265708754472
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40