bitscore colors: <40, 40-50 , 50-80, 80-200, >200
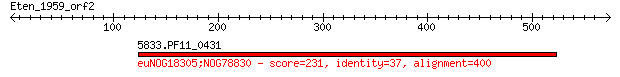
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: eggV2
2,483,276 sequences; 915,453,621 total letters
Query= Eten_1959_orf2
Length=574
Score E
Sequences producing significant alignments: (Bits) Value
5833.PF11_0431 231 6e-59
> 5833.PF11_0431
Length=504
Score = 231 bits (589), Expect = 6e-59, Method: Compositional matrix adjust.
Identities = 148/431 (34%), Positives = 235/431 (54%), Gaps = 33/431 (7%)
Query 123 EVPKFVPEYVEKIVEVPEVKVVDKIVELPRVEYQYKYVQKIETQENIIQRPKYETKYVEK 182
E K+ EY KI++VPE+K VDK+V P + + KYV K + NII++P + EK
Sbjct 71 EKSKYQFEYQNKIIQVPELKYVDKMVYDPVIIEKVKYVPKEVIKYNIIKKPVIKNIITEK 130
Query 183 IVEVPQVKEVVRYQDVEEVEEVIRYVPKGQVASEEWERLKEKHE--QEVKMEREAREAQY 240
V+V QVKE + +++ E VE+V YV K + +W + +E +++ ++ +
Sbjct 131 KVDVLQVKEKISFKEEEIVEDVYNYVDKD--LNTKWNESQYDNEMYKDLTKKKNYIGQNH 188
Query 241 KAEQEYQQLIAEEQAAYEAKRMGMPSTGPPPMP-----PAPRLEQVFKPKVVKTVEIQKH 295
++ Y+ + + P P +E VF P V K +E+ K
Sbjct 189 LLPNNINKMGHINDRTYKHINNNITNLLEPFGPQIDVEENKIIENVFVPNVEKVIEVNKK 248
Query 296 VPVSVDVPVPYMVPKPIVVPVQVPVLKFRDHFVPVPVRRRVVPRIRWTNEVYEVECIKEK 355
+ + +++PVPY+VPKP ++ V VPV KF D +VPVPV ++++P+I WT+++Y+V+C+ EK
Sbjct 249 IDIPINLPVPYIVPKPKIIDVDVPVFKFNDKYVPVPVSKKIIPKITWTDKIYQVDCLIEK 308
Query 356 PYLQVQDIIKPVPCDVEIKVHEFVEKAQPINPAELSQADVHAMWMRVNADL--------- 406
PYL +IIK VP D +I V E+ + + INP EL + D A+WMRVNADL
Sbjct 309 PYLVYHNIIKMVPTDSKITVREYPKGIKKINPEELYEVDNLALWMRVNADLKQEHDQMKN 368
Query 407 ----AEKRK-----EELGDKYPYVQHPEGTVFGEPGIKRTEE-----NAELSDPEAREGV 452
K+K E+L D E + E K + E N E + + E +
Sbjct 369 EKYETNKKKGKGETEQLDDNISSDHTCECSESYETYEKLSNEEFNSSNEETTIKSSNENI 428
Query 453 -EPLALHPGHPLNMTYLQNEWIKKETTATHEMYTPEWFEAHQKAMHNLTMQYPTQVQLSA 511
+ L LHPGHPL +LQN+WI ++TT +MY ++ +AH+ A+ NLT Q P + ++ A
Sbjct 429 LDTLPLHPGHPLEFIHLQNKWINQDTTNIPDMYDQKYLDAHRNAVFNLTTQMPREAEVEA 488
Query 512 EQAARMQQETQ 522
+Q +Q++ Q
Sbjct 489 KQLLYIQKKLQ 499
Lambda K H
0.313 0.129 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 256385640280
Database: eggV2
Posted date: Dec 15, 2009 4:47 PM
Number of letters in database: 915,453,621
Number of sequences in database: 2,483,276
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40