bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0027_orf1
Length=120
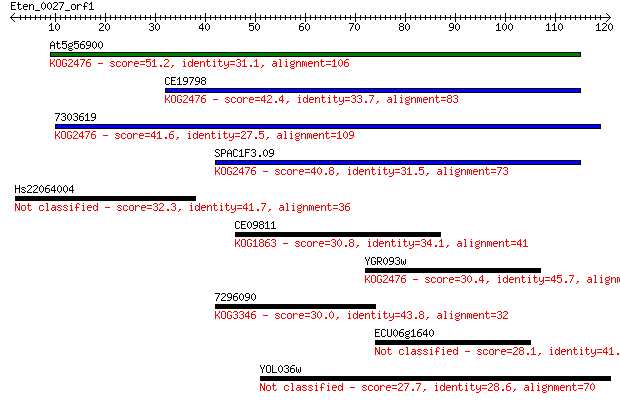
Score E
Sequences producing significant alignments: (Bits) Value
At5g56900 51.2 4e-07
CE19798 42.4 2e-04
7303619 41.6 3e-04
SPAC1F3.09 40.8 6e-04
Hs22064004 32.3 0.24
CE09811 30.8 0.70
YGR093w 30.4 0.83
7296090 30.0 1.2
ECU06g1640 28.1 3.8
YOL036w 27.7 4.9
> At5g56900
Length=593
Score = 51.2 bits (121), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 54/108 (50%), Gaps = 12/108 (11%)
Query 9 LFEKRAKRAGLSL--QKLPETADITSLSELANDPCLGYFYVEVPGVLTAKGQTIDRYLHV 66
+F A++ G L +K ++ D + + LG FYVE+P G + L
Sbjct 486 IFSLAAEKLGFKLVTKKFNDSTDGRKYLQKEYNAALGLFYVELP-----DGTVLSHTLE- 539
Query 67 QPGRDSGKIPMTFGREVMAELLGLRDKVNWQACLVPKDEEAKLAAALR 114
++ P FGREV+A LL + D+ +W+ C + ++EEAKLA +
Sbjct 540 ----ENEVFPAQFGREVLAGLLKIPDRADWRNCKISQEEEAKLAEDFK 583
> CE19798
Length=533
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/85 (32%), Positives = 44/85 (51%), Gaps = 15/85 (17%)
Query 32 SLSELANDPCLGYFYVEVPGVLTAKGQTIDRYLHVQPGRDSGKIPMTFGREVMAE--LLG 89
SL ++ N+ C YF E+P G + R P+ FGREV+A +L
Sbjct 447 SLLDMVNEGC-PYFVAELP-----DGSKLFT-------RSMKGFPLHFGREVLASTPILD 493
Query 90 LRDKVNWQACLVPKDEEAKLAAALR 114
DKV+W+AC++ K++E +L L+
Sbjct 494 CEDKVDWKACVLAKEKEVELVNKLK 518
> 7303619
Length=537
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 43/111 (38%), Gaps = 19/111 (17%)
Query 10 FEKRAKRAGLSLQKLPETADITSLSELANDPCLGYFYVEVPGVLTAKGQTIDRYLHVQPG 69
FE +A+ L + LP L E+ YF E+P T + + +
Sbjct 437 FEDKAEEFNLEFETLPALDSEKMLPEMG-----PYFLAELPDDSTLITRQMKHF------ 485
Query 70 RDSGKIPMTFGREVMA--ELLGLRDKVNWQACLVPKDEEAKLAAALRDVLG 118
P+ F R+V LL +KVNW+ CL+ KDEE R
Sbjct 486 ------PIHFARDVFCSENLLNCDEKVNWKDCLLDKDEEVAYVEDFRKAFA 530
> SPAC1F3.09
Length=561
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 38/75 (50%), Gaps = 14/75 (18%)
Query 42 LGYFYVEVPG--VLTAKGQTIDRYLHVQPGRDSGKIPMTFGREVMAELLGLRDKVNWQAC 99
L YF V +P +L + Q +R+ + FGR A++LGL D+V+W+ C
Sbjct 490 LNYFRVFLPSGKILIHRLQLRERF------------DLQFGRRAAAKILGLEDRVDWRKC 537
Query 100 LVPKDEEAKLAAALR 114
+ +DEE + A +
Sbjct 538 VQTEDEEKAESEAFK 552
> Hs22064004
Length=363
Score = 32.3 bits (72), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 23/36 (63%), Gaps = 0/36 (0%)
Query 2 EALQSGELFEKRAKRAGLSLQKLPETADITSLSELA 37
EA+QSGE+ A+R+G S QK P+ +T ++A
Sbjct 250 EAVQSGEMARSGAQRSGASTQKQPQCRALTPAGQVA 285
> CE09811
Length=1230
Score = 30.8 bits (68), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 24/41 (58%), Gaps = 3/41 (7%)
Query 46 YVEVPGVLTAKGQTIDRYLHVQPGRDSGKIPMTFGREVMAE 86
YV++P V+ + ++RY + GR K PM + RE+ A+
Sbjct 296 YVKLPPVIVIQ---LNRYKYTSKGRQKLKTPMAYPREIPAK 333
> YGR093w
Length=507
Score = 30.4 bits (67), Expect = 0.83, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 72 SGKIPMTFGREVMAELLGLRDKVNWQA--CLVPKDEE 106
S I + FGR V+A LL L +V W + CL K +E
Sbjct 450 SETIDLQFGRRVLAFLLNLPRRVKWNSSTCLQTKQQE 486
> 7296090
Length=210
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 14/32 (43%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 42 LGYFYVEVPGVLTAKGQTIDRYLHVQPGRDSG 73
L + V VPG+ KGQ I Y P +DSG
Sbjct 106 LHWLVVNVPGLDIMKGQPISEYFGPLPPKDSG 137
> ECU06g1640
Length=395
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 17/31 (54%), Gaps = 3/31 (9%)
Query 74 KIPMTFGREVMAELLGLRDKVNWQACLVPKD 104
KIP+ F V +LL R + NW C P+D
Sbjct 174 KIPLEF---VTTDLLDYRVEANWSLCNFPQD 201
> YOL036w
Length=761
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 36/70 (51%), Gaps = 1/70 (1%)
Query 51 GVLTAKGQTIDRYLHVQPGRDSGKIPMTFGREVMAELLGLRDKVNWQACLVPKDEEAKLA 110
GVL+ KG + + + P +G++ +T ++ E + RD ++ + L P D KLA
Sbjct 115 GVLSDKGNSKEELALLPPLPHTGEMEITPQFDI-NEAIFERDDISHSSRLEPDDVLTKLA 173
Query 111 AALRDVLGAN 120
+ RD G +
Sbjct 174 NSTRDATGED 183
Lambda K H
0.317 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40