bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0035_orf1
Length=285
Score E
Sequences producing significant alignments: (Bits) Value
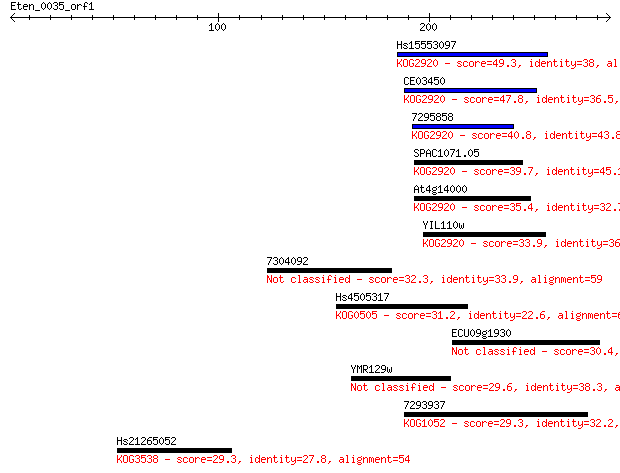
Hs15553097 49.3 8e-06
CE03450 47.8 3e-05
7295858 40.8 0.003
SPAC1071.05 39.7 0.007
At4g14000 35.4 0.13
YIL110w 33.9 0.40
7304092 32.3 1.3
Hs4505317 31.2 2.7
ECU09g1930 30.4 4.1
YMR129w 29.6 7.8
7293937 29.3 9.4
Hs21265052 29.3 9.8
> Hs15553097
Length=372
Score = 49.3 bits (116), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 27/73 (36%), Positives = 40/73 (54%), Gaps = 2/73 (2%)
Query 185 TTEIAGVKFSAILCSEGTYRQEAFEPLANVLEKFLHTHGVALVASKRYYFGVGGGSLAFM 244
++E VK+ IL SE Y + + L + L +G L+ASK +YFGVGGG F
Sbjct 279 SSEKLFVKYDLILTSETIYNPDYYSNLHQTFLRLLSKNGRVLLASKAHYFGVGGGVHLFQ 338
Query 245 AFLCEK--YSTRL 255
F+ E+ + TR+
Sbjct 339 KFVEERDVFKTRI 351
> CE03450
Length=229
Score = 47.8 bits (112), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 37/63 (58%), Gaps = 0/63 (0%)
Query 188 IAGVKFSAILCSEGTYRQEAFEPLANVLEKFLHTHGVALVASKRYYFGVGGGSLAFMAFL 247
++G KF IL SE Y ++ ++ L + + + GVA A+K +YFGVGG AF F+
Sbjct 142 LSGRKFDFILSSETIYNEDDYQALHDAVAATIKDDGVAWFAAKFFYFGVGGSVPAFCEFI 201
Query 248 CEK 250
++
Sbjct 202 NQR 204
> 7295858
Length=249
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/48 (43%), Positives = 27/48 (56%), Gaps = 0/48 (0%)
Query 192 KFSAILCSEGTYRQEAFEPLANVLEKFLHTHGVALVASKRYYFGVGGG 239
K+ IL SE Y + L + L + GV LVA+K +YFGVGGG
Sbjct 166 KYDLILTSETIYNIANQQKLLDTFAGRLKSDGVILVAAKSHYFGVGGG 213
> SPAC1071.05
Length=339
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query 193 FSAILCSEGTYRQEAFEPLANVLEKFLHTHGVALVASKRYYFGVGGGSLAF 243
FS +L SE Y + E +L K +T +ALVA K YFGVGG L F
Sbjct 239 FSLVLASETIYSLPSLENFLYMLLK--NTKNLALVAGKDLYFGVGGSILEF 287
> At4g14000
Length=269
Score = 35.4 bits (80), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 33/56 (58%), Gaps = 1/56 (1%)
Query 193 FSAILCSEGTYRQEAFEPLANVLEKFL-HTHGVALVASKRYYFGVGGGSLAFMAFL 247
+ IL +E Y A + ++++ L + G +A+K+YYFGVGGG+ F++ +
Sbjct 188 YDIILMAETIYSISAQKSQYELIKRCLAYPDGAVYMAAKKYYFGVGGGTRQFLSMI 243
> YIL110w
Length=377
Score = 33.9 bits (76), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 9/64 (14%)
Query 197 LCSEGTYRQEAFEPLANVLEKFLHTHGV------ALVASKRYYFGVGGGSLAFMAFLCEK 250
L SE Y+ + +A E L H + VA+K YFGVGG F A+L +K
Sbjct 288 LSSETIYQPDNLPVIA---ETILDIHNLPQTDVKTYVAAKDIYFGVGGSITEFEAYLDDK 344
Query 251 YSTR 254
++
Sbjct 345 INSE 348
> 7304092
Length=629
Score = 32.3 bits (72), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 27/59 (45%), Gaps = 5/59 (8%)
Query 123 SFCLLKGRALCLAADWEHFPKICCSCQGIVTGLKERKESETEEEQGAREHRQRETSVDK 181
S C LK + + + E P I G+VT KE+ET EEQ + H + DK
Sbjct 215 SRCTLKEGKMSESKNQETSPDILSEKSGVVT-----KENETREEQQDKTHLESNKIPDK 268
> Hs4505317
Length=1030
Score = 31.2 bits (69), Expect = 2.7, Method: Composition-based stats.
Identities = 14/63 (22%), Positives = 36/63 (57%), Gaps = 1/63 (1%)
Query 156 KERKESETEEEQGAREHRQRETSVDKTQKTTEIAGVKFSAILCSEGTYRQ-EAFEPLANV 214
+ +E +++ E+G+ + + S+ + + ++ AG ++ ++L G+Y E +P ++
Sbjct 864 ENEQEQQSDTEEGSNKKETQTDSISRYETSSTSAGDRYDSLLGRSGSYSYLEERKPYSSR 923
Query 215 LEK 217
LEK
Sbjct 924 LEK 926
> ECU09g1930
Length=440
Score = 30.4 bits (67), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 20/70 (28%), Positives = 31/70 (44%), Gaps = 6/70 (8%)
Query 211 LANVLEKFLHTHGVALVASKRYYFGVGGGSLAFMAFLCEKYSTRLSVEVAASFRSPNSSN 270
+ N+L L ++ K YF SL F+ F C + S+R V +S S
Sbjct 96 IINLLRHHLVDEVYSMYKDKPKYFSNHSNSLVFLVFFCAQSSSRRGVFGDSSLSS----- 150
Query 271 FRDILSVRIK 280
RD + +R+K
Sbjct 151 -RDFMFMRVK 159
> YMR129w
Length=1337
Score = 29.6 bits (65), Expect = 7.8, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Query 163 TEEEQGAREHRQRETSVDKTQKTTEIAGVKFSAILCSEGTYRQEAFE 209
TEE G +H +R + V +T K T+I ++ I +GTY EA E
Sbjct 1279 TEETDG--KHGKRRSQVVETHKVTDIYSHEYKVITSLQGTY--EAIE 1321
> 7293937
Length=885
Score = 29.3 bits (64), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 38/92 (41%), Gaps = 7/92 (7%)
Query 188 IAGVKFSAILCSEGTYRQEAFEPLANVLEKFLHTHGVALVASKRYYFGVGGGSLAFMA-- 245
AGV I E + Q + + ++L L + G A + GG LAF+A
Sbjct 238 FAGVLLWMIFHLERHWMQRCLDFIPSLLSSCLISFGAACIQGSYLMPKSAGGRLAFIAVM 297
Query 246 ---FLCEKYSTRLSVEVAASFRSPNSSNFRDI 274
FL Y T S+ V+ SP SN R I
Sbjct 298 LTSFLMYNYYT--SIVVSTLLGSPVRSNIRTI 327
> Hs21265052
Length=1223
Score = 29.3 bits (64), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 15/54 (27%), Positives = 22/54 (40%), Gaps = 0/54 (0%)
Query 52 KVLASVTRWNILKNTHDGLRTPAHQHQLFVSSSGFSPLAFAPSSEATEPACSVA 105
+ L V RW + D H H +F++ F P +AP + P S A
Sbjct 330 RSLEQVCRWAHSQQRQDPSHAEHHDHVVFLTRQDFGPSGYAPVTGMCHPLRSCA 383
Lambda K H
0.321 0.132 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6304412188
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40