bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0052_orf2
Length=150
Score E
Sequences producing significant alignments: (Bits) Value
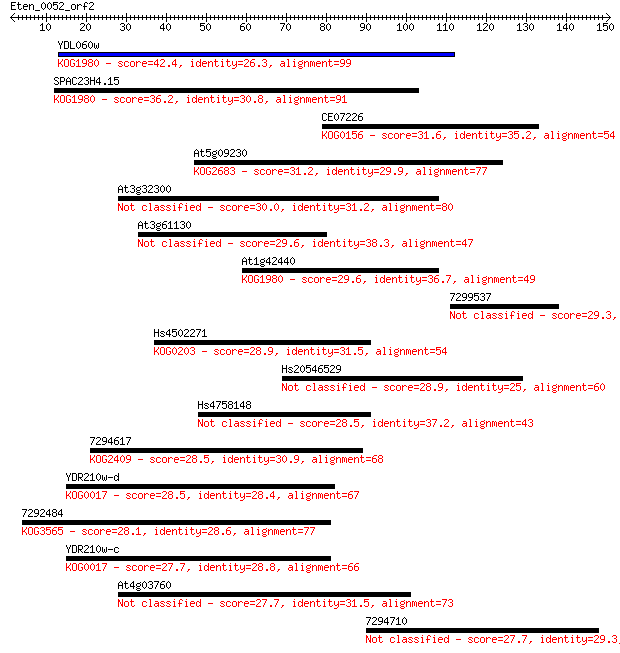
YDL060w 42.4 3e-04
SPAC23H4.15 36.2 0.023
CE07226 31.6 0.58
At5g09230 31.2 0.78
At3g32300 30.0 1.9
At3g61130 29.6 2.4
At1g42440 29.6 2.4
7299537 29.3 3.1
Hs4502271 28.9 3.8
Hs20546529 28.9 4.0
Hs4758148 28.5 5.0
7294617 28.5 5.1
YDR210w-d 28.5 5.3
7292484 28.1 6.9
YDR210w-c 27.7 8.5
At4g03760 27.7 8.6
7294710 27.7 9.3
> YDL060w
Length=788
Score = 42.4 bits (98), Expect = 3e-04, Method: Composition-based stats.
Identities = 26/102 (25%), Positives = 51/102 (50%), Gaps = 3/102 (2%)
Query 13 GHRHRSALKQQNKAFKGSSSKSKSLKE--KRRTAAANAASLAVNKPITKSERVQQSKQRR 70
GH HRS+LK +K++K + +LK K + + +K ++K +R ++KQ R
Sbjct 3 GHSHRSSLKNGHKSYKSKHASKGALKRLYKGKVEKEPVGTGKPDKQVSKLQRKNKAKQLR 62
Query 71 VQRARD-LHQRQVPSADGPPPKVVVLLPFSDSVCIDKVFHKL 111
QR D + R++ K++ ++P + + + +KL
Sbjct 63 AQRILDSIENRKLFEGKNGAAKIITIVPLVNDLDPLDILYKL 104
> SPAC23H4.15
Length=783
Score = 36.2 bits (82), Expect = 0.023, Method: Composition-based stats.
Identities = 28/95 (29%), Positives = 46/95 (48%), Gaps = 6/95 (6%)
Query 12 MGHRHRSALKQQNKAFKGSSSKSKSLKEKRRTAAANAASLAVN--KPITKSERVQQSKQR 69
M H HRS K + K FK + SLKEK + S N TK++R +KQ
Sbjct 1 MAHHHRSTFKAK-KPFKSKHASKSSLKEKYKNEVEPHRSGPKNIVHTSTKADRRNTAKQI 59
Query 70 RVQRARD--LHQRQVPSADGPPPKVVVLLPFSDSV 102
++ + + ++ R +G PKV+ ++P ++V
Sbjct 60 QLNKRTEVAMNNRIFGGKNG-APKVITIVPLCNNV 93
> CE07226
Length=236
Score = 31.6 bits (70), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 3/54 (5%)
Query 79 QRQVPSADGPPPKVVVLLPFSDSVCIDKVFHKLSQDLVQDHGKVFCEGIAGMDM 132
+R +P P P V LL ++ V K +KL L Q +G VF +AG+ M
Sbjct 22 RRNLPPGPAPLPFVGNLLMLTEKV---KPGYKLWDSLTQQYGSVFTFWMAGLPM 72
> At5g09230
Length=451
Score = 31.2 bits (69), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 23/85 (27%), Positives = 37/85 (43%), Gaps = 8/85 (9%)
Query 47 NAASLAVNKPITKSERVQQSKQRRVQRARDLHQRQVPSADGPPPKVVVL--------LPF 98
N A + KPIT E + S+ RR AR + +A P P L + F
Sbjct 192 NGAYSSGFKPITHQEFTRSSRARRRYWARSYAGWRRFTAAQPGPAHTALASLEKAGRINF 251
Query 99 SDSVCIDKVFHKLSQDLVQDHGKVF 123
+ +D++ H+ D ++ HG V+
Sbjct 252 MITQNVDRLHHRAGSDPLELHGTVY 276
> At3g32300
Length=567
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 25/81 (30%), Positives = 34/81 (41%), Gaps = 1/81 (1%)
Query 28 KGSSSKSKSLKEKRRTAAANAASLAVNKP-ITKSERVQQSKQRRVQRARDLHQRQVPSAD 86
K K + L E++R A A S V P T++ V V Q P+A
Sbjct 376 KKKEDKERRLAEEKRLADAGLISPRVASPDATQNRDVAPEVAAPVDPTPYKAQEVDPTAA 435
Query 87 GPPPKVVVLLPFSDSVCIDKV 107
P P+ VV LP SD+ +V
Sbjct 436 APLPEAVVALPASDNAAGKRV 456
> At3g61130
Length=673
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 26/47 (55%), Gaps = 1/47 (2%)
Query 33 KSKSLKEKRRTAAANAASLAVNKPITKSERVQQSKQRRVQRARDLHQ 79
KS +L EKR + + ++ V P TK R Q ++RR RA +L Q
Sbjct 129 KSNNLNEKRDSISKDSIHQKVETP-TKIHRRQLREKRREMRANELVQ 174
> At1g42440
Length=766
Score = 29.6 bits (65), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query 59 KSERVQQSKQRRVQ-RARDLHQRQVPSADGPPPKVVVLLPFSDSVCIDKV 107
K+ RVQ+ K R Q RA L +++ P+V+VL P S SV ++ +
Sbjct 20 KAARVQRGKMLREQKRAAVLKEKRASGGINSAPRVIVLFPLSASVELNSL 69
> 7299537
Length=581
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 19/30 (63%), Gaps = 3/30 (10%)
Query 111 LSQDLVQDHGKVFCEGIA---GMDMDGDDN 137
L+ ++V D GK F EGI GMD D D++
Sbjct 534 LNNEIVADQGKEFAEGIHLEDGMDEDQDED 563
> Hs4502271
Length=1020
Score = 28.9 bits (63), Expect = 3.8, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 30/54 (55%), Gaps = 1/54 (1%)
Query 37 LKEKRRTAAANAASLAVNKPITKSERVQQSKQRRVQRARDLHQRQVPSADGPPP 90
L E ++ A + L++++ + + +V SK QRA+D+ R P+A PPP
Sbjct 30 LDELKKEVAMDDHKLSLDE-LGRKYQVDLSKGLTNQRAQDVLARDGPNALTPPP 82
> Hs20546529
Length=253
Score = 28.9 bits (63), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 15/60 (25%), Positives = 30/60 (50%), Gaps = 7/60 (11%)
Query 69 RRVQRARDLHQRQVPSADGPPPKVVVLLPFSDSVCIDKVFHKLSQDLVQDHGKVFCEGIA 128
+R + R L + +VP+++ PP ++ L P + + D ++Q+ C+GIA
Sbjct 26 KRTGKGRSLRKPKVPASNLPPRQIPTLHPLAPTGSPDNELDNMTQEF-------HCQGIA 78
> Hs4758148
Length=331
Score = 28.5 bits (62), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 29/48 (60%), Gaps = 5/48 (10%)
Query 48 AASLAVNKPITKSERVQQSKQR----RVQRARDLHQ-RQVPSADGPPP 90
A ++A+N I K+E VQ++ +R R+Q+ + LH R + ++ PP
Sbjct 270 ALAVALNWDIKKTETVQEACERELALRLQQTQSLHSLRSISASKASPP 317
> 7294617
Length=855
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 31/68 (45%), Gaps = 4/68 (5%)
Query 21 KQQNKAFKGSSSKSKSLKEKRRTAAANAASLAVNKPITKSERVQQSKQRRVQRARDLHQR 80
K QN+A K + +KS + +A A A N P K QSK ++ Q +H+
Sbjct 726 KPQNQANKSAKTKSNQPFKTTESAPAKAEKSNGNNPFNKP----QSKSQQRQELPPIHKN 781
Query 81 QVPSADGP 88
Q + GP
Sbjct 782 QGGNKKGP 789
> YDR210w-d
Length=1755
Score = 28.5 bits (62), Expect = 5.3, Method: Composition-based stats.
Identities = 19/76 (25%), Positives = 37/76 (48%), Gaps = 9/76 (11%)
Query 15 RHRSALKQQNKAFKGSS-------SKSKSLKEKRRTAAANAASLAVNKPITKSERVQQSK 67
R+RS K ++++ ++ + K+ K +TA A+ S ++N P T ++ + +S
Sbjct 362 RNRSDEKNDSRSYTNTTKPKVIARNPQKTNNSKSKTARAHNVSTSINSPSTDNDSISKST 421
Query 68 QRRVQ--RARDLHQRQ 81
+Q DLH Q
Sbjct 422 TEPIQLNNKHDLHLGQ 437
> 7292484
Length=525
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 22/82 (26%), Positives = 41/82 (50%), Gaps = 9/82 (10%)
Query 4 VGDLAIVTMGHRHRSALKQQNKAFKGSSSKSKSLKEKRRTAAANAASLAVN-----KPIT 58
VGDLA G R + Q + G + SK+L+E+R+ ++ A+L N +
Sbjct 21 VGDLA----GQREVVSESLQLQIIAGVTLLSKTLREERKKCLSDGATLQQNLTTQLSSLD 76
Query 59 KSERVQQSKQRRVQRARDLHQR 80
+++R + R ++A D ++R
Sbjct 77 RAKRNYEKAYRDSEKAVDSYKR 98
> YDR210w-c
Length=440
Score = 27.7 bits (60), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 37/75 (49%), Gaps = 9/75 (12%)
Query 15 RHRSALKQQNKAFKGSS-------SKSKSLKEKRRTAAANAASLAVNKPITKSERVQQSK 67
R+RS K ++++ ++ + K+ K +TA A+ S ++N P T ++ + +S
Sbjct 362 RNRSDEKNDSRSYTNTTKPKVIARNPQKTNNSKSKTARAHNVSTSINSPSTDNDSISKST 421
Query 68 QRRVQ--RARDLHQR 80
+Q DLH R
Sbjct 422 TEPIQLNNKHDLHLR 436
> At4g03760
Length=723
Score = 27.7 bits (60), Expect = 8.6, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 30/74 (40%), Gaps = 1/74 (1%)
Query 28 KGSSSKSKSLKEKRRTAAANAASLAVNKP-ITKSERVQQSKQRRVQRARDLHQRQVPSAD 86
K K + L E++R A A V P T+ V V Q P+A
Sbjct 399 KKKEDKERRLAEEKRLANAGLILPRVASPDATQDRNVAPEVAAPVDPTPSEAQEVDPTAV 458
Query 87 GPPPKVVVLLPFSD 100
P P+ VV+LP SD
Sbjct 459 APLPEAVVVLPASD 472
> 7294710
Length=537
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Query 90 PKVVVLLPFSDSVCIDKVFHKLSQDLVQDHGKVFCEGIAGMDMDGDDNTSXDVVSPSI 147
PK L SD C+D + L DL Q+ K I+ +++G+ T+ D+++P +
Sbjct 367 PKTGNLTNVSDVSCLDTTYRLLCADL-QNKSKDNTGAIS-QNLEGNKTTAKDLITPDL 422
Lambda K H
0.315 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1852391706
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40