bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0096_orf4
Length=217
Score E
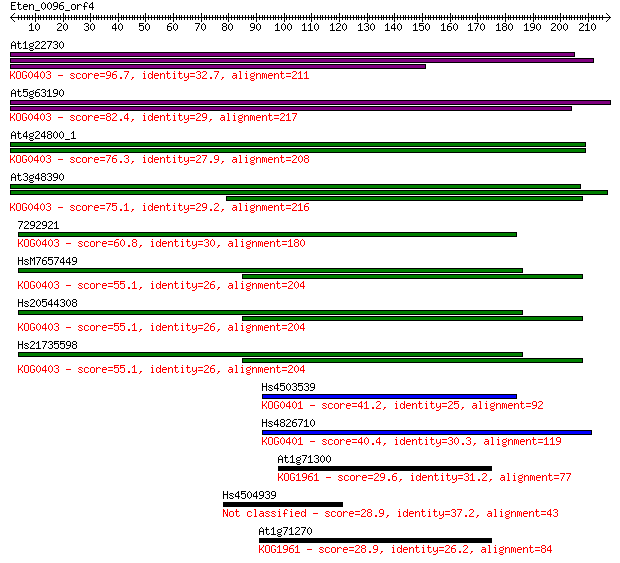
Sequences producing significant alignments: (Bits) Value
At1g22730 96.7 3e-20
At5g63190 82.4 6e-16
At4g24800_1 76.3 5e-14
At3g48390 75.1 1e-13
7292921 60.8 2e-09
HsM7657449 55.1 1e-07
Hs20544308 55.1 1e-07
Hs21735598 55.1 1e-07
Hs4503539 41.2 0.002
Hs4826710 40.4 0.003
At1g71300 29.6 5.0
Hs4504939 28.9 7.7
At1g71270 28.9 8.3
> At1g22730
Length=693
Score = 96.7 bits (239), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 65/204 (31%), Positives = 115/204 (56%), Gaps = 9/204 (4%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCE 60
+ DDL++D+P A D L +ARA+VD++LPPAFL + +L G++VL+KA+K
Sbjct 173 SADDLSVDIPDAVDVLAVFVARAIVDDILPPAFLKKQMKLLPDNSKGVEVLRKAEKSYLA 232
Query 61 QPGKAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQ 120
P A E K W GTD E K + +++Y+ + D EA + I + + P
Sbjct 233 TPLHA--EVVEKRWGGTDNWTAED--VKARINDLLKEYVMSGDKKEAFRCIKGLKV-PFF 287
Query 121 DSEVVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIEL 180
E+V++A I +MER K A ++ LL++ +G ++ + +G ++I D + D+ L
Sbjct 288 HHEIVKRALIMAMERRK----AQVRLLDLLKETIEVGLINSTQVTKGFSRIIDSIEDLSL 343
Query 181 DVPNAQQKICYMVAASRNKGLLPA 204
D+P+A++ + ++ + ++G L A
Sbjct 344 DIPDARRILQSFISKAASEGWLCA 367
Score = 61.2 bits (147), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 60/217 (27%), Positives = 104/217 (47%), Gaps = 16/217 (7%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGD--RYRLHFGGIGGMQVLKKAQKWL 58
+ DD ALD P + L LARAVVDE+L P L + G G +V++ A+ L
Sbjct 472 SADDTALDNPVVVEDLAMFLARAVVDEVLAPRDLEEVLNQTPEAGSSVGEKVIQMAKTLL 531
Query 59 CEQPGKAASERFRKIWTG----TDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDM 114
+ + ER + W G T+ + K +++ + +Y+ D EA++ + ++
Sbjct 532 ---KARLSGERILRCWGGGGIETNSPGSTVKEVKEKIQILLEEYVSGGDLREASRCVKEL 588
Query 115 NLAPDQDSEVVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDL 174
+ P EVV+K+ + +E ++ E+ K LL+ G + + +G ++ +
Sbjct 589 GM-PFFHHEVVKKSVVRIIEEKENEERLWK----LLKVCFDSGLVTIYQMTKGFKRVDES 643
Query 175 LPDIELDVPNAQQKICYMVAASRNKGLLPADYVFAEE 211
L D+ LDVP+A +K V + +G L D FA E
Sbjct 644 LEDLSLDVPDAAKKFSSCVERGKLEGFL--DESFASE 678
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 33/153 (21%), Positives = 63/153 (41%), Gaps = 37/153 (24%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCE 60
+++DL+LD+P A L +++A ++ WLC
Sbjct 337 SIEDLSLDIPDARRILQSFISKAA-----------------------------SEGWLCA 367
Query 61 QPGKAAS-ERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAI-IDMNLAP 118
K+ S + K+ + A +FK + K+ IR+Y + D +E + ++N +
Sbjct 368 SSLKSLSADAGEKLLENS-----SANVFKDKAKSIIREYFLSGDTSEVVHCLDTELNASS 422
Query 119 DQDSEVVRKAYI-FSMERSKDPEQAAKAVVGLL 150
Q + K I +M+R K ++ A +V L
Sbjct 423 SQLRAIFVKYLITLAMDRKKREKEMACVLVSTL 455
> At5g63190
Length=729
Score = 82.4 bits (202), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 63/217 (29%), Positives = 112/217 (51%), Gaps = 11/217 (5%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCE 60
+VDDLA+D+ A + L +ARA+VDE+LPP FL ++ G QV+ A+K
Sbjct 232 SVDDLAVDILDAVNVLALFIARAIVDEILPPVFLVRSKKILPESCKGFQVIVTAEKSYLS 291
Query 61 QPGKAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQ 120
P A E K W G+ T E K ++ +++Y++ D EA + I ++ ++
Sbjct 292 APHHA--ELVEKKWGGSTHTTVEET--KKKISEILKEYVENGDTYEACRCIRELGVSFFH 347
Query 121 DSEVVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIEL 180
EVV++A + +M D A V+ LL++ G + +++G ++ + L D+ L
Sbjct 348 -HEVVKRALVLAM----DSPTAESLVLKLLKETAEEGLISSSQMVKGFFRVAESLDDLAL 402
Query 181 DVPNAQQKICYMVAASRNKGLLPADYVFAEEAVEDGQ 217
D+P+A++ +V + + G L D F + +DG+
Sbjct 403 DIPSAKKLFDSIVPKAISGGWL--DDSFKITSDQDGE 437
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 58/204 (28%), Positives = 99/204 (48%), Gaps = 13/204 (6%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCE 60
+ +D ALD+ A + L LARAV+D++L P L D G + ++ A+ +
Sbjct 530 SAEDTALDIMDASNELALFLARAVIDDVLAPLNLEDISTKLPPKSTGTETVRSARSLIS- 588
Query 61 QPGKAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQ 120
+ A ER + W G + E K ++ + +Y +EA + I D+ + P
Sbjct 589 --ARHAGERLLRSWGGGTGWIVEDA--KDKISKLLEEYETGGVTSEACQCIRDLGM-PFF 643
Query 121 DSEVVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIEL 180
+ EVV+KA + +ME+ D ++ LLE+ G + + +G ++ D L D+ L
Sbjct 644 NHEVVKKALVMAMEKQND------RLLNLLEECFGEGLITTNQMTKGFGRVNDSLDDLSL 697
Query 181 DVPNAQQKI-CYMVAASRNKGLLP 203
D+PNA++K Y A N +LP
Sbjct 698 DIPNAKEKFELYASHAMDNGWILP 721
> At4g24800_1
Length=743
Score = 76.3 bits (186), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 58/209 (27%), Positives = 109/209 (52%), Gaps = 11/209 (5%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCE 60
+ DD +D+P A + L LARAVVD++LPPAFL + G QV++ A+K
Sbjct 199 SADDFVVDIPDAVNVLALFLARAVVDDILPPAFLPRAAKALPITSKGYQVVQTAEKSYLS 258
Query 61 QPGKAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQ 120
A E + W G T E K ++ + +Y++T + EA + + ++ ++
Sbjct 259 AAHHA--ELVERRWGGQTRTTVEE--VKKKIADILNEYVETGETYEACRCVRELGVSFFH 314
Query 121 DSEVVRKAYIFSMERSKDPEQAAKA-VVGLLEQLRALGELDQECIIQGLAQIKDLLPDIE 179
EVV++A + ++E AA+A V+ LL + + + +++G +++++ L D+
Sbjct 315 -HEVVKRALVTALEN-----HAAEAPVLKLLNEAASENLISSSQMVKGFSRLRESLDDLA 368
Query 180 LDVPNAQQKICYMVAASRNKGLLPADYVF 208
LD+P+A+ K +V + + G L A + +
Sbjct 369 LDIPSARTKFGLIVPKAVSGGWLDASFGY 397
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 62/213 (29%), Positives = 106/213 (49%), Gaps = 16/213 (7%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCE 60
+ +D ALD+ A + L LARAV+D++L P L + G + +K A+ +
Sbjct 497 SAEDTALDILDASNELALFLARAVIDDVLAPFNLEEISSKLRPNSSGTETVKMARSLIF- 555
Query 61 QPGKAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQ 120
+ A ER + W G E K ++ + +Y + +EA K I ++ + P
Sbjct 556 --ARHAGERLLRCWGGGSGWAVEDA--KDKISNLLEEYESSGLVSEACKCIHELGM-PFF 610
Query 121 DSEVVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIEL 180
+ EVV+KA + ME+ KD K ++ LL++ + G + + +G ++KD L D+ L
Sbjct 611 NHEVVKKALVMGMEKKKD-----KMMLDLLQESFSEGLITTNQMTKGFTRVKDGLEDLAL 665
Query 181 DVPNAQQKIC-YMVAASRN----KGLLPADYVF 208
D+PNA++K Y+ +N KG L A+ F
Sbjct 666 DIPNAKEKFNDYVEYGKKNETTRKGSLEANMKF 698
> At3g48390
Length=633
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 60/207 (28%), Positives = 104/207 (50%), Gaps = 14/207 (6%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCE 60
+ +D ALD+ A D L LARAV+D++L P L + G + ++ A+ +
Sbjct 434 SAEDTALDILAASDELALFLARAVIDDVLAPLNLEEISNSLPPKSTGSETIRSARSLIS- 492
Query 61 QPGKAASERFRKIWTG-TDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPD 119
+ A ER + W G T + +A K ++ + +Y +EA + I D+ + P
Sbjct 493 --ARHAGERLLRSWGGGTGWAVEDA---KDKIWKLLEEYEVGGVISEACRCIRDLGM-PF 546
Query 120 QDSEVVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIE 179
+ EVV+KA + +ME+ D ++ LL++ A G + + +G ++KD L D+
Sbjct 547 FNHEVVKKALVMAMEKKND------RMLNLLQECFAEGIITTNQMTKGFGRVKDSLDDLS 600
Query 180 LDVPNAQQKICYMVAASRNKGLLPADY 206
LD+PNA++K VA + G L D+
Sbjct 601 LDIPNAEEKFNSYVAHAEENGWLHRDF 627
Score = 74.7 bits (182), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 61/218 (27%), Positives = 107/218 (49%), Gaps = 14/218 (6%)
Query 1 AVDDLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCE 60
+V DLALD+P A + L +ARA+VDE+LPP FL + G QV+ ++
Sbjct 139 SVGDLALDIPDAVNVLALFIARAIVDEILPPVFLARAKKTLPHSSQGFQVILVSENSYLS 198
Query 61 QPGKAASERFRKIWTG-TDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPD 119
P A E W G T T+ E K ++ + +Y++ D EA + I ++ ++
Sbjct 199 APHHA--ELVETKWGGSTHITVEET---KRKISEFLNEYVENGDTREACRCIRELGVSFF 253
Query 120 QDSEVVRKAYIFSME-RSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDI 178
E+V+ + ME R+ +P ++ LL++ G + + +G +++ D L D+
Sbjct 254 H-HEIVKSGLVLVMESRTSEP-----LILKLLKEATEEGLISSSQMAKGFSRVADSLDDL 307
Query 179 ELDVPNAQQKICYMVAASRNKGLLPADYVFAEEAVEDG 216
LD+P+A+ +V + G L D F E + ++G
Sbjct 308 SLDIPSAKTLFESIVPKAIIGGWLDEDS-FKERSDQNG 344
Score = 33.5 bits (75), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 29/129 (22%), Positives = 61/129 (47%), Gaps = 5/129 (3%)
Query 79 PTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQDSEVVRKAYIFSMERSKD 138
P +K E+ + I +Y + D AA ++D+ L+ + V++ +M+R
Sbjct 49 PVFNPLEDYKREVVSIIDEYFSSGDVEVAASDLMDLGLS-EYHPYFVKRLVSMAMDRGNK 107
Query 139 PEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIELDVPNAQQKICYMVAASRN 198
++ A LL +L AL + + I G ++ + + D+ LD+P+A + +A +
Sbjct 108 EKEKASV---LLSRLYAL-VVSPDQIRVGFIRLLESVGDLALDIPDAVNVLALFIARAIV 163
Query 199 KGLLPADYV 207
+LP ++
Sbjct 164 DEILPPVFL 172
> 7292921
Length=507
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 54/190 (28%), Positives = 90/190 (47%), Gaps = 25/190 (13%)
Query 4 DLALDVPQAHDFLTKSLARAVVDELLPPAFL---GDRYRLHFG-GIGGMQVLKKAQKWLC 59
DL LD P+A L +ARAV D+ +PP F+ G+ R H G G Q L++A +
Sbjct 282 DLVLDTPEAPIMLGNFMARAVADDCIPPKFVAKSGEELR-HLGLGEHAEQALRRADSLIY 340
Query 60 EQPGKAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPD 119
K +W G +R + +++ +++YL + D AEA + + + + P
Sbjct 341 ----KHVWAHLDNVW-GMGGPLRPVKTITMQMELLLKEYLSSRDVAEAQRCLRALEV-PH 394
Query 120 QDSEVVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECII------QGLAQIKD 173
E+V +A + ++E + +A+ LL+Q LD C++ QG + D
Sbjct 395 YHHELVYEAIVMTLESLS--QTTEEAMCELLKQ------LDLTCLVLPAGMEQGFLRAFD 446
Query 174 LLPDIELDVP 183
+ DI LDVP
Sbjct 447 DMADIVLDVP 456
> HsM7657449
Length=469
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 48/182 (26%), Positives = 88/182 (48%), Gaps = 8/182 (4%)
Query 4 DLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCEQPG 63
+LALD P+A + + +ARAV D +L ++ D Y+ + L KA L G
Sbjct 249 ELALDTPRAPQLVGQFIARAVGDGILCNTYI-DSYKGTVDCVQARAALDKATVLLSMSKG 307
Query 64 KAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQDSE 123
+R +W G+ + E+ +++YL + D +EA + ++ + P E
Sbjct 308 ---GKRKDSVW-GSGGGQQSVNHLVKEIDMLLKEYLLSGDISEAEHCLKELEV-PHFHHE 362
Query 124 VVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIELDVP 183
+V +A I +E + E K ++ LL+ L + + + +G +I + +PDI LDVP
Sbjct 363 LVYEAIIMVLESTG--ESTFKMILDLLKSLWKSSTITVDQMKRGYERIYNEIPDINLDVP 420
Query 184 NA 185
++
Sbjct 421 HS 422
Score = 33.1 bits (74), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 29/124 (23%), Positives = 58/124 (46%), Gaps = 7/124 (5%)
Query 85 RIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQDSEVVRKAYIFSME-RSKDPEQAA 143
R F+ L I++Y + D E A+ + D+NL + S V A ++E ++ E +
Sbjct 162 RAFEKTLTPIIQEYFEHGDTNEVAEMLRDLNLG-EMKSGVPVLAVSLALEGKASHREMTS 220
Query 144 KAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIELDVPNAQQKICYMVAASRNKGLLP 203
K + L + + ++++ ++ LP++ LD P A Q + +A + G+L
Sbjct 221 KFLSDLCGTVMSTTDVEKS-----FDKLLKDLPELALDTPRAPQLVGQFIARAVGDGILC 275
Query 204 ADYV 207
Y+
Sbjct 276 NTYI 279
> Hs20544308
Length=469
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 48/182 (26%), Positives = 88/182 (48%), Gaps = 8/182 (4%)
Query 4 DLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCEQPG 63
+LALD P+A + + +ARAV D +L ++ D Y+ + L KA L G
Sbjct 249 ELALDTPRAPQLVGQFIARAVGDGILCNTYI-DSYKGTVDCVQARAALDKATVLLSMSKG 307
Query 64 KAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQDSE 123
+R +W G+ + E+ +++YL + D +EA + ++ + P E
Sbjct 308 ---GKRKDSVW-GSGGGQQSVNHLVKEIDMLLKEYLLSGDISEAEHCLKELEV-PHFHHE 362
Query 124 VVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIELDVP 183
+V +A I +E + E K ++ LL+ L + + + +G +I + +PDI LDVP
Sbjct 363 LVYEAIIMVLESTG--ESTFKMILDLLKSLWKSSTITVDQMKRGYERIYNEIPDINLDVP 420
Query 184 NA 185
++
Sbjct 421 HS 422
Score = 32.7 bits (73), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 58/124 (46%), Gaps = 7/124 (5%)
Query 85 RIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQDSEVVRKAYIFSME-RSKDPEQAA 143
R F+ L I++Y + D E A+ + D+NL + S V A ++E ++ E +
Sbjct 162 RAFEKTLTPIIQEYFEHGDTNEVAEMLRDLNLG-EMKSGVPVLAVSLALEGKASHREMTS 220
Query 144 KAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIELDVPNAQQKICYMVAASRNKGLLP 203
K + L + + D E L +KDL P++ LD P A Q + +A + G+L
Sbjct 221 KLLSDLCGTV--MSTTDVEKSFDKL--LKDL-PELALDTPRAPQLVGQFIARAVGDGILC 275
Query 204 ADYV 207
Y+
Sbjct 276 NTYI 279
> Hs21735598
Length=458
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 48/182 (26%), Positives = 88/182 (48%), Gaps = 8/182 (4%)
Query 4 DLALDVPQAHDFLTKSLARAVVDELLPPAFLGDRYRLHFGGIGGMQVLKKAQKWLCEQPG 63
+LALD P+A + + +ARAV D +L ++ D Y+ + L KA L G
Sbjct 238 ELALDTPRAPQLVGQFIARAVGDGILCNTYI-DSYKGTVDCVQARAALDKATVLLSMSKG 296
Query 64 KAASERFRKIWTGTDPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQDSE 123
+R +W G+ + E+ +++YL + D +EA + ++ + P E
Sbjct 297 ---GKRKDSVW-GSGGGQQSVNHLVKEIDMLLKEYLLSGDISEAEHCLKELEV-PHFHHE 351
Query 124 VVRKAYIFSMERSKDPEQAAKAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIELDVP 183
+V +A I +E + E K ++ LL+ L + + + +G +I + +PDI LDVP
Sbjct 352 LVYEAIIMVLESTG--ESTFKMILDLLKSLWKSSTITVDQMKRGYERIYNEIPDINLDVP 409
Query 184 NA 185
++
Sbjct 410 HS 411
Score = 33.1 bits (74), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 58/124 (46%), Gaps = 7/124 (5%)
Query 85 RIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQDSEVVRKAYIFSME-RSKDPEQAA 143
R F+ L I++Y + D E A+ + D+NL + S V A ++E ++ E +
Sbjct 151 RAFEKTLTPIIQEYFEHGDTNEVAEMLRDLNLG-EMKSGVPVLAVSLALEGKASHREMTS 209
Query 144 KAVVGLLEQLRALGELDQECIIQGLAQIKDLLPDIELDVPNAQQKICYMVAASRNKGLLP 203
K + L + + D E L +KDL P++ LD P A Q + +A + G+L
Sbjct 210 KLLSDLCGTV--MSTTDVEKSFDKL--LKDL-PELALDTPRAPQLVGQFIARAVGDGILC 264
Query 204 ADYV 207
Y+
Sbjct 265 NTYI 268
> Hs4503539
Length=907
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/92 (25%), Positives = 48/92 (52%), Gaps = 3/92 (3%)
Query 92 KACIRKYLDTLDAAEAAKAIIDMNLAPDQDSEVVRKAYIFSMERSKDPEQAAKAVVGLLE 151
+ + +YL++ +A EA + +M E++ K I S++RS + ++ A +++ LL+
Sbjct 549 ETVVTEYLNSGNANEAVNGVREMRAPKHFLPEMLSKVIILSLDRSDEDKEKASSLISLLK 608
Query 152 QLRALGELDQECIIQGLAQIKDLLPDIELDVP 183
Q G + +Q + D P +E+D+P
Sbjct 609 Q---EGIATSDNFMQAFLNVLDQCPKLEVDIP 637
> Hs4826710
Length=1396
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 36/120 (30%), Positives = 57/120 (47%), Gaps = 6/120 (5%)
Query 92 KACIRKYLDTLDAAEAAKAIIDMNLAPDQDSEVVRKAYIFSMERSKDPEQAAKAVVG-LL 150
KA I +YL D EA + + ++ +P VR ++ERS A+ +G LL
Sbjct 1049 KAIIEEYLHLNDMKEAVQCVQEL-ASPSLLFIFVRHGVESTLERSA----IAREHMGQLL 1103
Query 151 EQLRALGELDQECIIQGLAQIKDLLPDIELDVPNAQQKICYMVAASRNKGLLPADYVFAE 210
QL G L QGL +I +L D+E+D+P+ + +V +G +P +F E
Sbjct 1104 HQLLCAGHLSTAQYYQGLYEILELAEDMEIDIPHVWLYLAELVTPILQEGGVPMGELFRE 1163
> At1g71300
Length=701
Score = 29.6 bits (65), Expect = 5.0, Method: Composition-based stats.
Identities = 24/87 (27%), Positives = 42/87 (48%), Gaps = 12/87 (13%)
Query 98 YLDTLDAAEAA----------KAIIDMNLAPDQDSEVVRKAYIFSMERSKDPEQAAKAVV 147
Y+DT++ +A K +D+ + D R +FS RSK+P + AV
Sbjct 275 YIDTMNKVLSAHFQSYIQAFEKLQLDIATSNDLIGVDTRSTGLFS--RSKEPLKNRCAVF 332
Query 148 GLLEQLRALGELDQECIIQGLAQIKDL 174
L E+++ + E+DQ +I +A+ L
Sbjct 333 ALGERIQIIKEIDQPALIPHIAEASSL 359
> Hs4504939
Length=1227
Score = 28.9 bits (63), Expect = 7.7, Method: Composition-based stats.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 78 DPTMREARIFKHELKACIRKYLDTLDAAEAAKAIIDMNLAPDQ 120
D M+EA+ KH+ A + K+ L+ AEAA + I+ A +Q
Sbjct 978 DYFMQEAKRMKHKADAMVEKFGKALNYAEAALSFIECGNAMEQ 1020
> At1g71270
Length=735
Score = 28.9 bits (63), Expect = 8.3, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 41/84 (48%), Gaps = 7/84 (8%)
Query 91 LKACIRKYLDTLDAAEAAKAIIDMNLAPDQDSEVVRKAYIFSMERSKDPEQAAKAVVGLL 150
L A R Y+ L+ K +D+ A D R +FS R+++P + AV L
Sbjct 317 LSAHFRAYIQALE-----KLQLDIATAYDLIGVETRTTGLFS--RAREPLKNRSAVFALG 369
Query 151 EQLRALGELDQECIIQGLAQIKDL 174
++++ + ++DQ +I +A+ L
Sbjct 370 DRIKIIKDIDQPALIPHIAEASSL 393
Lambda K H
0.320 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4040519078
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40