bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0128_orf1
Length=305
Score E
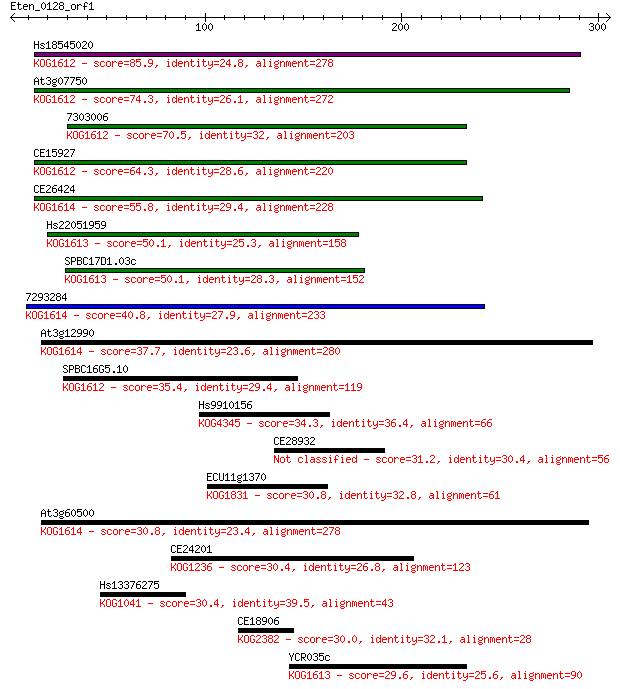
Sequences producing significant alignments: (Bits) Value
Hs18545020 85.9 1e-16
At3g07750 74.3 3e-13
7303006 70.5 4e-12
CE15927 64.3 3e-10
CE26424 55.8 1e-07
Hs22051959 50.1 6e-06
SPBC17D1.03c 50.1 6e-06
7293284 40.8 0.004
At3g12990 37.7 0.031
SPBC16G5.10 35.4 0.13
Hs9910156 34.3 0.36
CE28932 31.2 2.8
ECU11g1370 30.8 3.3
At3g60500 30.8 3.4
CE24201 30.4 4.8
Hs13376275 30.4 5.2
CE18906 30.0 7.2
YCR035c 29.6 8.6
> Hs18545020
Length=291
Score = 85.9 bits (211), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 69/281 (24%), Positives = 127/281 (45%), Gaps = 10/281 (3%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDAD 72
+S AE ++ GV +R DGRG + R + D++ + SA ++ ++ V A+
Sbjct 6 LSEAEKVYIVHGVQEDLRVDGRGCEDYRCVEVETDVVSNTSGSARVKLGHTDILVGVKAE 65
Query 73 VILPGGGGNQEPY-SVSVGCGAAEQELADVTGGD--GSSLSSLLQAMLEQLLLPHLKPLT 129
+ P E Y V C A+ + GGD G+ +++ L + LK L
Sbjct 66 MGTPKLEKPNEGYLEFFVDCSASATPEFEGRGGDDLGTEIANTLYRIFNNKSSVDLKTLC 125
Query 130 DPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEY 189
W + +DV++L GG L DA+S+A+ AL ++P V V E++E + + +++
Sbjct 126 ISPREHCWVLYVDVLLLECGGNLFDAISIAVKAALFNTRIPRVRVLEDEEGSKDIELSDD 185
Query 190 KVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDSFLVAAFLPSGACVG 249
C ++ VP ++T +IG +V D T E C + L+ + G
Sbjct 186 PYDC-------IRLSVENVPCIVTLCKIGYRHVVDATLQEEACSLASLLVSVTSKGVVTC 238
Query 250 LQKFGHVLADCRTLQTTLTTSQRLSREILEDLQRQAEMKKS 290
++K G D ++ + T +R+ + + LQ ++S
Sbjct 239 MRKVGKGSLDPESIFEMMETGKRVGKVLHASLQSVVHKEES 279
> At3g07750
Length=293
Score = 74.3 bits (181), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 71/289 (24%), Positives = 129/289 (44%), Gaps = 26/289 (8%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDAD 72
+S E F++ G+A +R DGR R RP + ++P + SA ++ VIA+V A+
Sbjct 3 LSLGEQHFIKGGIAQDLRTDGRKRLTYRPIYVETGVIPQANGSARVRIGGTDVIASVKAE 62
Query 73 VILPGG-GGNQEPYSVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLP-------- 123
+ P ++ +V + C + GG+ LSS L L++ LL
Sbjct 63 IGRPSSLQPDKGKVAVFIDCSPTAEPTFGGRGGE--ELSSELALALQRCLLGGKSGAAIH 120
Query 124 --------HLKPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVE 175
+L L +G++ W + ID +V+ + G LLDA+ AI AL +P V V
Sbjct 121 VLVPGAGINLSSLLIKEGKVCWDLYIDGLVISSDGNLLDALGAAIKAALTNTAIPKVNVS 180
Query 176 EEDEKDQGLAITEYKVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATCGDS 235
E D+ E +S +E + + +VP+++T ++G Y+ D T E + S
Sbjct 181 AEVADDEQ---PEIDISDEEYLQ----FDTSSVPVIVTLTKVGTHYIVDATAEEESQMSS 233
Query 236 FLVAAFLPSGACVGLQKFGHVLADCRTLQTTLTTSQRLSREILEDLQRQ 284
+ + +G GL K D + ++ ++ ++ ++ L +
Sbjct 234 AVSISVNRTGHICGLTKRSGSGLDPSVILDMISVAKHVTETLMSKLDSE 282
> 7303006
Length=281
Score = 70.5 bits (171), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 65/209 (31%), Positives = 92/209 (44%), Gaps = 16/209 (7%)
Query 30 RGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGG----GGNQEPY 85
R DGR R + RP L L+ + SA ++ ++ V ++ +P G E +
Sbjct 8 RCDGRSRRDYRPMDLETGLVSNASGSARLRLANTDILVGVKTEIDVPNPLTPEFGKLEFF 67
Query 86 SVSVGCGA-AEQELADVTGGD-GSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQWRISIDV 143
V C A A E G D L LQ E L + + L G+ W++ ID+
Sbjct 68 ---VDCSANATPEFEGRGGSDLAQELILSLQNAYESPLAFNYRTLCLIPGQQCWKLYIDI 124
Query 144 IVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYKVSCDERIAAGTPY 203
++L GG L DAVSLA AL KLP V D L I++ C T
Sbjct 125 LILECGGNLHDAVSLAAKAALFNTKLPRVTATMLDAGITDLIISDNPYDC-------TRI 177
Query 204 PIDTVPILITAAQIGDTYVWDLTEIEATC 232
I+TVP+L+T +IGD + D + E C
Sbjct 178 GIETVPLLVTVCKIGDYVLVDPSAEEEVC 206
> CE15927
Length=305
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 63/230 (27%), Positives = 105/230 (45%), Gaps = 16/230 (6%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENT-VIATVDA 71
+S E F+ G +R DGR + R +L +L + S +Q + T V+A +
Sbjct 5 LSDDEKLFLITGAEANIRNDGRSCQDFRQIILERGVLSGTNGSCRVQLGQTTDVLAGIKL 64
Query 72 DVILPGGGGNQEP-----YSVSVGCGAAEQELADVTGGD--GSSLSSLLQAMLEQLL--L 122
++ +EP ++V A+ Q GGD LS+ QA + L +
Sbjct 65 ELESYDAETEKEPKQPINFNVDFSANASSQFAG--KGGDEYAEELSAAFQAAYSKALDIM 122
Query 123 PHLKPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQ 182
P+L DG +W+I++D+ VL+ G + DA+S+AI GAL L+ P V +D
Sbjct 123 PNLAKTQLADG-YRWKINVDISVLQWDGSVSDAISIAIIGALSDLEFPEGDVAPDDGGKV 181
Query 183 GLAITEYKVSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATC 232
+ + KV+ + R + P+L+T ++IG + D T E C
Sbjct 182 RIVLKRPKVAGNVR---DENVQVAICPLLLTISKIGTANLVDCTPEEECC 228
> CE26424
Length=434
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 67/237 (28%), Positives = 96/237 (40%), Gaps = 27/237 (11%)
Query 13 ISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENT-VIATVDA 71
++ E A + E + G R D R E R V L+ + AI T NT V+A V A
Sbjct 6 VTNCEKAVILEALKIGKRFDFRSLEEFRD----VKLVVGAEVGTAICTIGNTKVMAAVSA 61
Query 72 DVILPGGGGNQEPYS------VSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHL 125
++ P P+ V + A D G G L LL+ ++ +
Sbjct 62 EIAEPSS---MRPHKGVINIDVDLSPMANYANEHDRLGSKGMELIRLLELIIRDSRCIDV 118
Query 126 KPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLA 185
+ L G+ W+I +DV +L G LLD LA AL+ + PNV +E L
Sbjct 119 EALCIRAGKEIWKIRVDVRILDEDGSLLDCACLAAITALQHFRRPNVTLEPH----HTLI 174
Query 186 ITEYKVSCDERIAAGTPYPIDTVPILITAAQI--GDTYVWDLTEIEATCGDSFLVAA 240
+EY+ A P I +PI T + G V D T+ E C D +V A
Sbjct 175 YSEYE-------KAPVPLNIYHMPICTTIGLLDKGQIVVIDPTDKETACLDGSIVVA 224
> Hs22051959
Length=276
Score = 50.1 bits (118), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 40/161 (24%), Positives = 70/161 (43%), Gaps = 3/161 (1%)
Query 20 FVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGGG 79
+ R + R DGR E R + + + + SA ++ TVI V A+ P
Sbjct 13 YYRRFLKENCRPDGRELGEFRTTTVNIGSISTADGSALVKLGNTTVICGVKAEFAAPSTD 72
Query 80 GNQEPY---SVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQ 136
+ Y +V + + + + G + S + ++E + + L G+L
Sbjct 73 APDKGYVVPNVDLPPLCSSRFRSGPPGEEAQVASQFIADVIENSQIIQKEDLCISPGKLV 132
Query 137 WRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEE 177
W + D+I L G +LDA + A+ ALK ++LP V + EE
Sbjct 133 WVLYCDLICLDYDGNILDACTFALLAALKNVQLPEVTINEE 173
> SPBC17D1.03c
Length=270
Score = 50.1 bits (118), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 43/161 (26%), Positives = 71/161 (44%), Gaps = 15/161 (9%)
Query 29 VRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILPGGGGNQEPYSV- 87
VR DGR +E R V+ + + + SA I+ EN + + A++ P E + V
Sbjct 34 VRSDGRSVSEFREIVINDNCISTANGSAIIRAGENVFVCGIKAEIAEPFENSPNEGWIVP 93
Query 88 ----SVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLL----LPHLKPLTDPDGRLQWRI 139
S C + G S L+ ++ L Q L L +L+ L + + W +
Sbjct 94 NLELSPLCSSK------FKPGPPSDLAQVVSQELHQTLQQSNLINLQSLCIFEKKAAWVL 147
Query 140 SIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEK 180
D+I L G D A++ ALK +KLP + +E+ E+
Sbjct 148 YADIICLNYDGSAFDYAWAALFAALKTVKLPTAVWDEDLER 188
> 7293284
Length=466
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 65/246 (26%), Positives = 96/246 (39%), Gaps = 35/246 (14%)
Query 9 FQGAISRAETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIAT 68
F+ S+ E +FV+ V R DGR E R L S A+ + V+A
Sbjct 62 FEPGKSKPERSFVQLAVKQNQRLDGRRSNESRDVELTFGK---DWGSVAVSMGDTKVLAQ 118
Query 69 VDADVILPGGGGNQE---PYSVSVGCGAAEQELADVTGGDGS-SLSSLLQAMLEQLLLPH 124
V ++ P E +V +G G A + A T S L+SLL+
Sbjct 119 VTCEMGPPALSRPNEGKIHLNVYLG-GVAFLDEAHTTHDQRSLKLNSLLERTFRSSRSID 177
Query 125 LKPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGL 184
L+ L + W I ++V VL G L DA ++A AL + P+V KD L
Sbjct 178 LESLCVAVEKHVWCIRVNVNVLNHDGNLYDASTIATLAALMHFRRPDVWY-----KDGEL 232
Query 185 AITEYKVSCDERIAAGTPYPIDTVPILITAAQIGDTY---------VWDLTEIEATCGDS 235
I + K ER + +P+L + TY + D T +E DS
Sbjct 233 RIFKKK----ER---------EFIPLLFHHYPVSVTYCVYKSSVQPILDPTLLEENAADS 279
Query 236 FLVAAF 241
+V +F
Sbjct 280 VIVLSF 285
> At3g12990
Length=307
Score = 37.7 bits (86), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 66/295 (22%), Positives = 114/295 (38%), Gaps = 29/295 (9%)
Query 17 ETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILP 76
E+ FV + +R DGRG + R + S+ +Q + V+A V A ++ P
Sbjct 15 ESKFVESALQSELRVDGRGLYDYRKLTIKFGK---EYGSSQVQLGQTHVMAFVTAQLVQP 71
Query 77 GGGGNQEPYSVSVGCGAAEQELADVT---GGDGSS---LSSLLQAMLEQLLLPHLKPLTD 130
P S +AD + G G S L ++ L + + L
Sbjct 72 Y---KDRPSEGSFSIFTEFSPMADPSFEPGHPGESAVELGRIIDRALRESRAVDTESLCV 128
Query 131 PDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYK 190
G+L W + ID+ +L GG L+DA ++A AL + P+ V ++ +D + E +
Sbjct 129 LAGKLVWSVRIDLHILDNGGNLVDAANVAALAALMTFRRPDCTVGGDNSQDVIIHPPEER 188
Query 191 VSCDERIAAGTPYPIDTVPILITAA--QIGDTYVWDLTEIEATCGDSFLVAAFLPSGACV 248
P I +PI T G V D T +E + +G
Sbjct 189 --------EPLPLIIHHLPIAFTFGFFNKGSILVMDPTYVEEAVMCGRMTVTVNANGDIC 240
Query 249 GLQKFGH------VLADC-RTLQTTLTTSQRLSREILEDLQRQAEMKKSRLNGNL 296
+QK G V+ C R + + + ++ R+ +E R+ +K + + L
Sbjct 241 AIQKPGEEGVNQSVILHCLRLASSRASATTKIIRDAVEAYNRERSSQKVKRHHTL 295
> SPBC16G5.10
Length=299
Score = 35.4 bits (80), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 35/122 (28%), Positives = 56/122 (45%), Gaps = 11/122 (9%)
Query 28 GVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENT-VIATVDADV--ILPGGGGNQEP 84
+R DGR +LRP VD+LP + SA ++ + ++ V A+V P GG
Sbjct 21 AIRNDGRSIDQLRPLSGQVDVLPGTNGSARVKWASSVEIVIGVKAEVGDATPEGGKYVAS 80
Query 85 YSVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQWRISIDVI 144
+S +E ++ S L+S LQ +L L + +LK T W I +D +
Sbjct 81 VEISPSVSIQNRETDEIP----SFLTSALQDLLNALAVDYLK-FTPSKA---WIIHVDAV 132
Query 145 VL 146
V+
Sbjct 133 VI 134
> Hs9910156
Length=858
Score = 34.3 bits (77), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 36/74 (48%), Gaps = 12/74 (16%)
Query 97 ELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQWRISIDVIVLR--------A 148
+L D+T + S + + ++EQ +L L+ GRL W +S+D R
Sbjct 151 QLPDLTVYNEDFRSFIERDLIEQSMLVALEQA----GRLNWWVSVDPTSQRLLPLATTGD 206
Query 149 GGCLLDAVSLAIWG 162
G CLL A SL +WG
Sbjct 207 GNCLLHAASLGMWG 220
> CE28932
Length=277
Score = 31.2 bits (69), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 30/56 (53%), Gaps = 0/56 (0%)
Query 135 LQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYK 190
L+W++ I V VL G LDA A+ AL KLP++++ + + + T+ +
Sbjct 124 LEWQLHIIVKVLSLEGNFLDAAVCALGAALVDAKLPSIVLNHSEGDESTIEKTQIQ 179
> ECU11g1370
Length=1461
Score = 30.8 bits (68), Expect = 3.3, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 8/61 (13%)
Query 101 VTGGDGSSLSSLLQAMLEQLLLPHLKPLTDPDGRLQWRISIDVIVLRAGGCLLDAVSLAI 160
V +G +S LL LE +LP + L DP + D+IVL + + D ++L I
Sbjct 254 VLAKEGIPVSRLLDIFLEHKMLPLVSELLDPP-----ELCFDIIVLSS---IRDHLNLGI 305
Query 161 W 161
W
Sbjct 306 W 306
> At3g60500
Length=438
Score = 30.8 bits (68), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 65/302 (21%), Positives = 117/302 (38%), Gaps = 38/302 (12%)
Query 17 ETAFVREGVACGVRGDGRGRAELRPRVLAVDLLPFSCSSAAIQTEENTVIATVDADVILP 76
E+ FV + +R DGRG + R + S+ +Q + V+ V A ++ P
Sbjct 15 ESKFVETALQSELRVDGRGLYDYRKLTIK---FGKEYGSSEVQLGQTHVMGFVTAQLVQP 71
Query 77 GGGGNQEPYSVSVGCGAAEQELADVT------GGDGSSLSSLLQAMLEQLLLPHLKPLTD 130
P S+ +AD + G L ++ L + + L
Sbjct 72 Y---KDRPNEGSLSIFTEFSPMADPSFEPGRPGESAVELGRIIDRGLRESRAVDTESLCV 128
Query 131 PDGRLQWRISIDVIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAITEYK 190
G++ W + ID+ +L GG L+DA ++A AL + P+ V E+ G + +
Sbjct 129 LAGKMVWSVRIDLHILDNGGNLVDAANIAALAALMTFRRPDCTVGGEN----GQEVIIHP 184
Query 191 VSCDERIAAGTPYPIDTVPILITAA--QIGDTYVWDLTEIEATCGDSFLVAAFLPSGACV 248
+ E + P I +PI T G+ V D T +E + +G
Sbjct 185 LEEREPL----PLIIHHLPIAFTFGFFNKGNIVVMDPTYVEEAVMCGRMTVTVNANGDIC 240
Query 249 GLQKFGH------VLADCRTLQTT--LTTSQRLSREI--------LEDLQRQAEMKKSRL 292
+QK G V+ C L ++ T++ + E+ L+ ++R + KS +
Sbjct 241 AIQKPGEEGVNQSVILHCLRLASSRAAATTKIIREEVEAYNCERSLQKVKRHPTLAKSEV 300
Query 293 NG 294
+G
Sbjct 301 SG 302
> CE24201
Length=600
Score = 30.4 bits (67), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 33/135 (24%), Positives = 63/135 (46%), Gaps = 24/135 (17%)
Query 83 EPYSVSVGCGAAEQELADVTGGDGSSLSSLLQAMLEQLLLPHLKPLTD-------PDGRL 135
EP+S+ GC +A V G +S L +A ++ P LK T D +
Sbjct 198 EPFSIGSGC------IAQVYRGT-VDISELEKATGKEY--PELKGRTTQRIAIKVADKDV 248
Query 136 QWRISIDVIVLRAGGCLLDAVSLAIW-----GALKALKLPNVMVEEEDEKDQGLAITEYK 190
+I +D+ +LR+G L+ + A+W GAL+ ++ V+ + D +++ A+ ++
Sbjct 249 DKQIELDLSILRSGAWLMQHIVPALWYLDPTGALEQFEM--VLRRQVDLRNEAKALKKFS 306
Query 191 VSCDERIAAGTPYPI 205
+ + G +PI
Sbjct 307 DNFSSK-ETGIKFPI 320
> Hs13376275
Length=860
Score = 30.4 bits (67), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 22/45 (48%), Gaps = 2/45 (4%)
Query 47 DLLPFSCSSAAIQTEENTVIAT--VDADVILPGGGGNQEPYSVSV 89
D P ++ T +AT VD DV LPG GG P+ VS+
Sbjct 81 DRRPVYDGKRSLYTANPLPVATTGVDLDVTLPGEGGKDRPFKVSI 125
> CE18906
Length=299
Score = 30.0 bits (66), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 9/28 (32%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 117 LEQLLLPHLKPLTDPDGRLQWRISIDVI 144
+ Q +L +L+P ++ +G+++W+I+I+ I
Sbjct 195 MRQFILTNLQPSSENEGQMEWKININTI 222
> YCR035c
Length=394
Score = 29.6 bits (65), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 23/102 (22%), Positives = 44/102 (43%), Gaps = 24/102 (23%)
Query 143 VIVLRAGGCLLDAVSLAIWGALKALKLPNVMVEEEDEKDQGLAIT------------EYK 190
++VL G + D ++ AL+++KLP + DE+ L +T Y+
Sbjct 217 IVVLSRTGPVFDLCWNSLMYALQSVKLPRAFI---DERASDLRMTIRTRGRSATIRETYE 273
Query 191 VSCDERIAAGTPYPIDTVPILITAAQIGDTYVWDLTEIEATC 232
+ CD+ +VP++I A I + + E++ C
Sbjct 274 IICDQ---------TKSVPLMINAKNIAFASNYGIVELDPEC 306
Lambda K H
0.318 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6955109822
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40