bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0209_orf2
Length=116
Score E
Sequences producing significant alignments: (Bits) Value
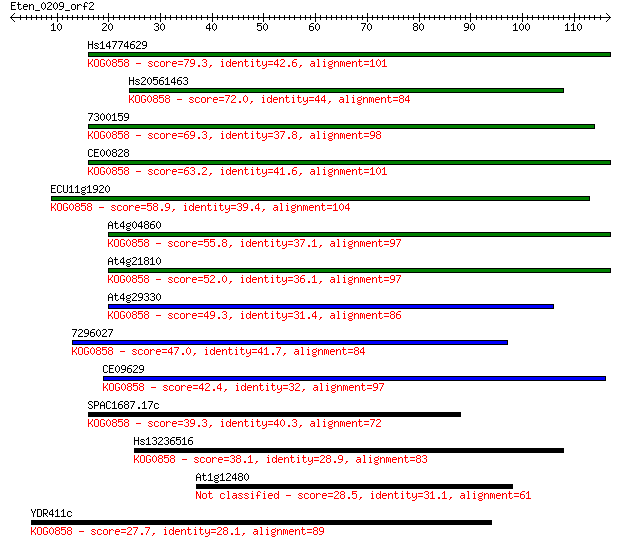
Hs14774629 79.3 1e-15
Hs20561463 72.0 2e-13
7300159 69.3 2e-12
CE00828 63.2 1e-10
ECU11g1920 58.9 2e-09
At4g04860 55.8 2e-08
At4g21810 52.0 3e-07
At4g29330 49.3 2e-06
7296027 47.0 8e-06
CE09629 42.4 2e-04
SPAC1687.17c 39.3 0.002
Hs13236516 38.1 0.004
At1g12480 28.5 3.2
YDR411c 27.7 6.1
> Hs14774629
Length=239
Score = 79.3 bits (194), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 43/102 (42%), Positives = 63/102 (61%), Gaps = 3/102 (2%)
Query 16 VSSYFFSGSMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLLVGWNMADHLV 74
VS F + ++ Y+W RRNP R++ F + QAP+LP++L SLL+G ++ L+
Sbjct 115 VSLVFLGQAFTIMLVYVWSRRNPYVRMNFFGLLNFQAPFLPWVLMGFSLLLGNSIIVDLL 174
Query 75 GIAVGHFYYFFEDVYPLLPTSKGFRIFRTPRILMWLLKQPED 116
GIAVGH Y+F EDV+P P G RI +TP IL + P++
Sbjct 175 GIAVGHIYFFLEDVFPNQP--GGIRILKTPSILKAIFDTPDE 214
> Hs20561463
Length=239
Score = 72.0 bits (175), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 37/85 (43%), Positives = 56/85 (65%), Gaps = 3/85 (3%)
Query 24 SMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLLVGWNMADHLVGIAVGHFY 82
+++ ++ Y+W RR+P R++ F + QAP+LP+ L SLL+G ++ L+GIAVGH Y
Sbjct 123 ALMAMLVYVWSRRSPRVRVNFFGLLTFQAPFLPWALMGFSLLLGNSILVDLLGIAVGHIY 182
Query 83 YFFEDVYPLLPTSKGFRIFRTPRIL 107
YF EDV+P P K R+ +TP L
Sbjct 183 YFLEDVFPNQPGGK--RLLQTPGFL 205
> 7300159
Length=261
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/99 (37%), Positives = 58/99 (58%), Gaps = 3/99 (3%)
Query 16 VSSYFFSGSMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLLVGWNMADHLV 74
V+ F + ++ Y+W RRNP ++ F + QAPYLP++L S+++G + ++
Sbjct 113 VNLLFLGQAFTLMLVYVWSRRNPLVPMNFFGVLNFQAPYLPWVLLCCSMILGNTVWVDVI 172
Query 75 GIAVGHFYYFFEDVYPLLPTSKGFRIFRTPRILMWLLKQ 113
G+ VGH YY EDVYP L S G+R+ +TP L L +
Sbjct 173 GMGVGHIYYVLEDVYPTL--SNGYRLIKTPYFLKRLFNE 209
> CE00828
Length=227
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/108 (38%), Positives = 60/108 (55%), Gaps = 9/108 (8%)
Query 16 VSSYFFSGSMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLLVGWNMADHLV 74
V F + ++ YIW RRNP +++ F + APYLP++L L SLL+G N +
Sbjct 103 VQILFLGQAFTIMLVYIWSRRNPMIQMNFFGVLTFTAPYLPWVLLLFSLLLGNNAVVDFM 162
Query 75 GIAVGHFYYFFEDVYPLLPTSKGFRIFRTPRILMWLLKQ------PED 116
GIA GH Y+F EDV+P G R +TP+ L++L + PED
Sbjct 163 GIACGHIYFFLEDVFPF--QEHGKRFLKTPQWLVYLFDERRPEPLPED 208
> ECU11g1920
Length=348
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 41/106 (38%), Positives = 59/106 (55%), Gaps = 11/106 (10%)
Query 9 AFSYIFGVSSYFFSGSMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLLVGW 67
A S I+G+S+ S +TYIW +RNP + IF F+ A YLPF+L LL
Sbjct 113 AISNIYGISA--LGTSFSATITYIWTKRNPRAIVQIFGFISFPAFYLPFILPGFMLLSRR 170
Query 68 NMA-DHLVGIAVGHFYYFFEDVYPLLPTSKGFRIFRTPRILMWLLK 112
+++ D ++GI VGH +++F+DVYP G I TP W+ K
Sbjct 171 SISIDDVLGIVVGHLFHYFKDVYPRW----GRDILSTP---CWVKK 209
> At4g04860
Length=244
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/105 (34%), Positives = 53/105 (50%), Gaps = 11/105 (10%)
Query 20 FFSGSMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLLVGWNMADHLVGIAV 78
F S S+ +M Y+W ++NP +S A YLP++L S+LVG + L+G+
Sbjct 129 FLSNSLTFMMVYVWSKQNPYIHMSFLGLFTFTAAYLPWVLLGFSILVGASAWVDLLGMIA 188
Query 79 GHFYYFFEDVYPLLPTSKGFRIFRTPRILMWL-------LKQPED 116
GH YYF +VYP + R +TP L L + +PED
Sbjct 189 GHAYYFLAEVYPRMTNR---RPLKTPSFLKALFADEPVVVARPED 230
> At4g21810
Length=225
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/105 (33%), Positives = 51/105 (48%), Gaps = 11/105 (10%)
Query 20 FFSGSMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLLVGWNMADHLVGIAV 78
F S S+ +M Y+W ++NP +S A YLP++L S+LVG + +G+
Sbjct 110 FLSNSLTFMMVYVWSKQNPYIHMSFLGLFTFTAAYLPWVLLGFSILVGASAWGDFLGMIA 169
Query 79 GHFYYFFEDVYPLLPTSKGFRIFRTPRILMWL-------LKQPED 116
GH YYF VYP + R +TP L L + +PED
Sbjct 170 GHAYYFLAFVYPRMTDR---RPLKTPSFLKALFADEPVVIARPED 211
> At4g29330
Length=281
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 49/87 (56%), Gaps = 4/87 (4%)
Query 20 FFSGSMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLLVGWNMADHLVGIAV 78
F S++ ++ Y+W R PN +S++ + ++A YLP+ + L ++ G + L+GI
Sbjct 134 FLGVSLVFMLLYLWSREFPNANISLYGLVTLKAFYLPWAMLALDVIFGSPIMPDLLGIIA 193
Query 79 GHFYYFFEDVYPLLPTSKGFRIFRTPR 105
GH YYF ++PL + G +TP+
Sbjct 194 GHLYYFLTVLHPL---ATGKNYLKTPK 217
> 7296027
Length=245
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 35/92 (38%), Positives = 52/92 (56%), Gaps = 11/92 (11%)
Query 13 IFGVSSYFFSGSMINVMTYIWGRRNPNTRLSIFF-MPVQAPYLPFLLALLSLLVGWNMAD 71
IF V YF +++ +TYIW + N + +S +F +A YLP++LA + +++A
Sbjct 114 IFNV--YFLMDTLVLAITYIWCQLNKDVTVSFWFGTRFKAMYLPWVLAAFEFIFHFSLAS 171
Query 72 HLVGIAVGHFYYFFEDVY-------PLLPTSK 96
LVGI VGH YYFF+ Y PLL T +
Sbjct 172 -LVGIFVGHVYYFFKFQYSQDLGGTPLLETPQ 202
> CE09629
Length=245
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 31/98 (31%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Query 19 YFFSGSMINVMTYIWGRRNPNTRLSIFF-MPVQAPYLPFLLALLSLLVGWNMADHLVGIA 77
YF M+ + Y+W + N +T +S +F M A YLP++L + ++ + LVGI
Sbjct 118 YFLLEPMVISVLYVWCQVNKDTIVSFWFGMRFPARYLPWVLWGFNAVLRGGGTNELVGIL 177
Query 78 VGHFYYFFEDVYPLLPTSKGFRIFRTPRILMWLLKQPE 115
VGH Y+F Y P G + TP L L+ +
Sbjct 178 VGHAYFFVALKY---PDEYGVDLISTPEFLHRLIPDED 212
> SPAC1687.17c
Length=168
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/79 (36%), Positives = 43/79 (54%), Gaps = 7/79 (8%)
Query 16 VSSYF----FSGSMIN-VMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLALLSLL-VGWN 68
V+SYF F+ S + M YIW ++P R+SI V+APY+P+++ LL L G
Sbjct 86 VTSYFSYMPFAASYFSFTMLYIWSWKHPLYRISILGLFDVKAPYVPWVMVLLRWLRTGIF 145
Query 69 MADHLVGIAVGHFYYFFED 87
L+ +GH Y+F D
Sbjct 146 PLLDLISALIGHVYFFVTD 164
> Hs13236516
Length=251
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 46/84 (54%), Gaps = 3/84 (3%)
Query 25 MINVMTYIWGRRNPNTRLSIFF-MPVQAPYLPFLLALLSLLVGWNMADHLVGIAVGHFYY 83
+I + Y+W + N + +S +F +A YLP+++ + ++G ++ + L+G VGH Y+
Sbjct 125 LIMSVLYVWAQLNRDMIVSFWFGTRFKACYLPWVILGFNYIIGGSVINELIGNLVGHLYF 184
Query 84 FFEDVYPLLPTSKGFRIFRTPRIL 107
F YP+ + F TP+ L
Sbjct 185 FLMFRYPMDLGGRNF--LSTPQFL 206
> At1g12480
Length=556
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 31/62 (50%), Gaps = 9/62 (14%)
Query 37 NPNTRLSIFFMPVQAPYLPFLLALLSLLVGWN-MADHLVGIAVGHFYYFFEDVYPLLPTS 95
NP++ LS+ F+ A+L+ VGW+ +A L + H+ F +Y LPTS
Sbjct 328 NPSSHLSVVGN--------FVGAILASKVGWDEVAKFLWAVGFAHYLVVFVTLYQRLPTS 379
Query 96 KG 97
+
Sbjct 380 EA 381
> YDR411c
Length=341
Score = 27.7 bits (60), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 25/96 (26%), Positives = 43/96 (44%), Gaps = 12/96 (12%)
Query 5 CVFQAFSYIFGVSSYF---FSGSMINVMTYIWGRRNPNTRLSIF-FMPVQAPYLPFLLAL 60
+ A + I+G SY+ + I+ +TY W N N ++ + +PV Y P +
Sbjct 133 AITTATTIIYG--SYYPVVLTSGFISCITYTWSIDNANVQIMFYGLIPVWGKYFPLIQLF 190
Query 61 LSLLVGWNMAD---HLVGIAVGHFYYFFEDVYPLLP 93
+S + +N D L+G G+ Y D + L P
Sbjct 191 ISFV--FNEGDFVISLIGFTTGYLYTCL-DTHTLGP 223
Lambda K H
0.334 0.147 0.494
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174970866
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40