bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0234_orf3
Length=136
Score E
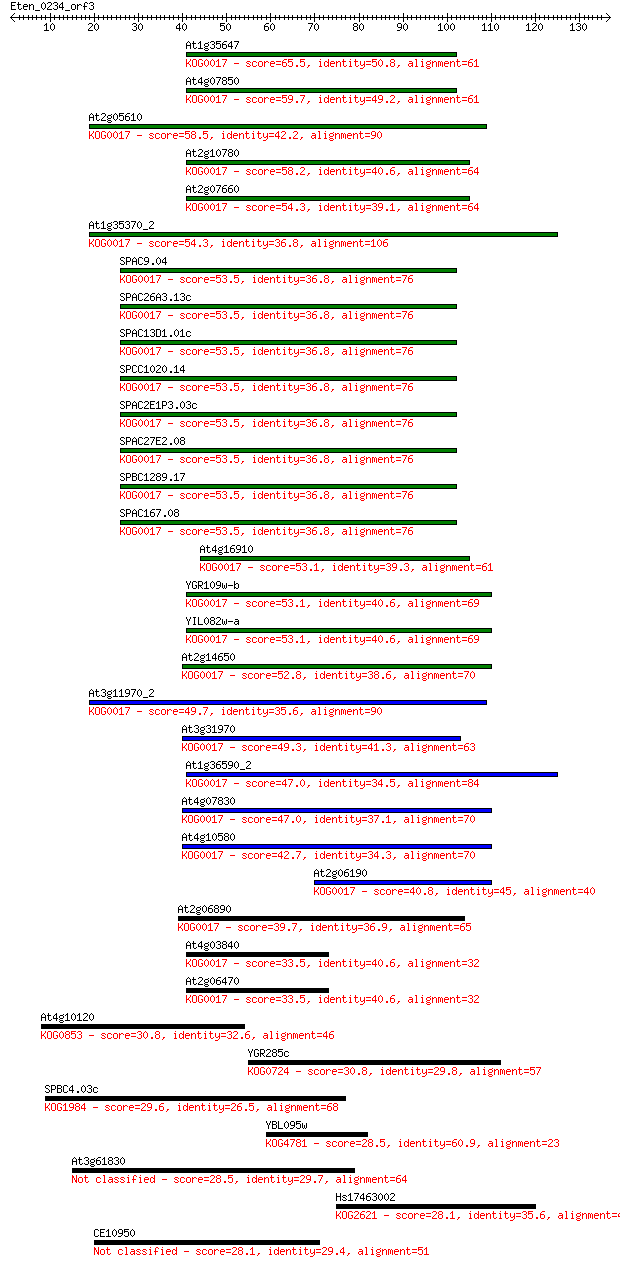
Sequences producing significant alignments: (Bits) Value
At1g35647 65.5 3e-11
At4g07850 59.7 2e-09
At2g05610 58.5 3e-09
At2g10780 58.2 5e-09
At2g07660 54.3 6e-08
At1g35370_2 54.3 7e-08
SPAC9.04 53.5 1e-07
SPAC26A3.13c 53.5 1e-07
SPAC13D1.01c 53.5 1e-07
SPCC1020.14 53.5 1e-07
SPAC2E1P3.03c 53.5 1e-07
SPAC27E2.08 53.5 1e-07
SPBC1289.17 53.5 1e-07
SPAC167.08 53.5 1e-07
At4g16910 53.1 1e-07
YGR109w-b 53.1 1e-07
YIL082w-a 53.1 2e-07
At2g14650 52.8 2e-07
At3g11970_2 49.7 2e-06
At3g31970 49.3 2e-06
At1g36590_2 47.0 1e-05
At4g07830 47.0 1e-05
At4g10580 42.7 2e-04
At2g06190 40.8 8e-04
At2g06890 39.7 0.002
At4g03840 33.5 0.12
At2g06470 33.5 0.12
At4g10120 30.8 0.69
YGR285c 30.8 0.83
SPBC4.03c 29.6 1.6
YBL095w 28.5 3.4
At3g61830 28.5 4.0
Hs17463002 28.1 4.5
CE10950 28.1 4.5
> At1g35647
Length=1495
Score = 65.5 bits (158), Expect = 3e-11, Method: Composition-based stats.
Identities = 31/61 (50%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHFV 100
C+ +K+ +Q P GL+ L IPS +S+DFV +P T TG DSI V+VD SKMAHF+
Sbjct 1040 CKQAKAKSQ-PHGLYTPLPIPSHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFI 1098
Query 101 P 101
P
Sbjct 1099 P 1099
> At4g07850
Length=1138
Score = 59.7 bits (143), Expect = 2e-09, Method: Composition-based stats.
Identities = 30/61 (49%), Positives = 39/61 (63%), Gaps = 1/61 (1%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHFV 100
C+ +K+ +Q P GL L IP +S+DFV +P T TG DSI V+VD SKMAHF+
Sbjct 807 CKQAKAKSQ-PHGLCTPLPIPLHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFI 865
Query 101 P 101
P
Sbjct 866 P 866
> At2g05610
Length=780
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 38/94 (40%), Positives = 51/94 (54%), Gaps = 5/94 (5%)
Query 19 HIRVHGL--WRICVPQFPEFLTH--VCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVT 74
H RV GL W+ V F+ C+ KS N GL Q L IP R + VS+DF+
Sbjct 494 HQRVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPIPDRIWSDVSMDFI- 552
Query 75 DIPPTTTGHDSILVMVDSLSKMAHFVPIANLTNA 108
D P + G I+V+VD LSK AHF+ +A+ +A
Sbjct 553 DGLPLSNGKTVIMVVVDRLSKAAHFIALAHPYSA 586
> At2g10780
Length=1611
Score = 58.2 bits (139), Expect = 5e-09, Method: Composition-based stats.
Identities = 26/65 (40%), Positives = 44/65 (67%), Gaps = 1/65 (1%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPP-TTTGHDSILVMVDSLSKMAHF 99
C+ K+ +Q P+GL Q L IP + H+++DFVT +P + H+++ V+VD L+K AHF
Sbjct 1208 CQLVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHF 1267
Query 100 VPIAN 104
+ I++
Sbjct 1268 MAISD 1272
> At2g07660
Length=949
Score = 54.3 bits (129), Expect = 6e-08, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 43/65 (66%), Gaps = 1/65 (1%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPP-TTTGHDSILVMVDSLSKMAHF 99
C+ K+ +Q P+GL Q L I + H+++DFVT +P + H+++ V+VD L+K AHF
Sbjct 600 CQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSAHF 659
Query 100 VPIAN 104
+ I++
Sbjct 660 MAISD 664
> At1g35370_2
Length=923
Score = 54.3 bits (129), Expect = 7e-08, Method: Composition-based stats.
Identities = 39/110 (35%), Positives = 53/110 (48%), Gaps = 16/110 (14%)
Query 19 HIRVHGL--WRICVPQFPEFLTH--VCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVT 74
H RV L W+ V F+ C+ KS N GL Q L IP + VS+DF+
Sbjct 550 HQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPLPIPDKIWCDVSMDFIE 609
Query 75 DIPPTTTGHDSILVMVDSLSKMAHFVPIANLTNASGSVCFRPWNLLTTQQ 124
+P + G I+V+VD LSK AHFV +A+ P++ LT Q
Sbjct 610 GLP-NSGGKSVIMVVVDRLSKAAHFVALAH-----------PYSALTVAQ 647
> SPAC9.04
Length=1333
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 26 WRICVPQFPEFLT--HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGH 83
W+ Q E++ H C+ +KS N KP G Q + R +S+DF+T + P ++G+
Sbjct 943 WKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL-PESSGY 1001
Query 84 DSILVMVDSLSKMAHFVP 101
+++ V+VD SKMA VP
Sbjct 1002 NALFVVVDRFSKMAILVP 1019
> SPAC26A3.13c
Length=1333
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 26 WRICVPQFPEFLT--HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGH 83
W+ Q E++ H C+ +KS N KP G Q + R +S+DF+T + P ++G+
Sbjct 943 WKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL-PESSGY 1001
Query 84 DSILVMVDSLSKMAHFVP 101
+++ V+VD SKMA VP
Sbjct 1002 NALFVVVDRFSKMAILVP 1019
> SPAC13D1.01c
Length=1333
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 26 WRICVPQFPEFLT--HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGH 83
W+ Q E++ H C+ +KS N KP G Q + R +S+DF+T + P ++G+
Sbjct 943 WKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL-PESSGY 1001
Query 84 DSILVMVDSLSKMAHFVP 101
+++ V+VD SKMA VP
Sbjct 1002 NALFVVVDRFSKMAILVP 1019
> SPCC1020.14
Length=1333
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 26 WRICVPQFPEFLT--HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGH 83
W+ Q E++ H C+ +KS N KP G Q + R +S+DF+T + P ++G+
Sbjct 943 WKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL-PESSGY 1001
Query 84 DSILVMVDSLSKMAHFVP 101
+++ V+VD SKMA VP
Sbjct 1002 NALFVVVDRFSKMAILVP 1019
> SPAC2E1P3.03c
Length=1333
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 26 WRICVPQFPEFLT--HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGH 83
W+ Q E++ H C+ +KS N KP G Q + R +S+DF+T + P ++G+
Sbjct 943 WKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL-PESSGY 1001
Query 84 DSILVMVDSLSKMAHFVP 101
+++ V+VD SKMA VP
Sbjct 1002 NALFVVVDRFSKMAILVP 1019
> SPAC27E2.08
Length=1333
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 26 WRICVPQFPEFLT--HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGH 83
W+ Q E++ H C+ +KS N KP G Q + R +S+DF+T + P ++G+
Sbjct 943 WKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL-PESSGY 1001
Query 84 DSILVMVDSLSKMAHFVP 101
+++ V+VD SKMA VP
Sbjct 1002 NALFVVVDRFSKMAILVP 1019
> SPBC1289.17
Length=1333
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 26 WRICVPQFPEFLT--HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGH 83
W+ Q E++ H C+ +KS N KP G Q + R +S+DF+T + P ++G+
Sbjct 943 WKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL-PESSGY 1001
Query 84 DSILVMVDSLSKMAHFVP 101
+++ V+VD SKMA VP
Sbjct 1002 NALFVVVDRFSKMAILVP 1019
> SPAC167.08
Length=1214
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/78 (35%), Positives = 45/78 (57%), Gaps = 3/78 (3%)
Query 26 WRICVPQFPEFLT--HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGH 83
W+ Q E++ H C+ +KS N KP G Q + R +S+DF+T + P ++G+
Sbjct 824 WKGIRKQIQEYVQNCHTCQINKSRNHKPYGPLQPIPPSERPWESLSMDFITAL-PESSGY 882
Query 84 DSILVMVDSLSKMAHFVP 101
+++ V+VD SKMA VP
Sbjct 883 NALFVVVDRFSKMAILVP 900
> At4g16910
Length=687
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 24/62 (38%), Positives = 41/62 (66%), Gaps = 1/62 (1%)
Query 44 SKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPP-TTTGHDSILVMVDSLSKMAHFVPI 102
+K +Q P+G+ Q L IP + H+ +DFVT +P + H+++ V+VD L+K AHF+ I
Sbjct 295 AKEEHQVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAI 354
Query 103 AN 104
++
Sbjct 355 SD 356
> YGR109w-b
Length=1547
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/69 (40%), Positives = 39/69 (56%), Gaps = 0/69 (0%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHFV 100
C+ KS + GL Q L I R +S+DFVT +PPT+ + ILV+VD SK AHF+
Sbjct 1145 CQLIKSHRPRLHGLLQPLPIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFI 1204
Query 101 PIANLTNAS 109
+A+
Sbjct 1205 ATRKTLDAT 1213
> YIL082w-a
Length=1498
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 28/69 (40%), Positives = 39/69 (56%), Gaps = 0/69 (0%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHFV 100
C+ KS + GL Q L I R +S+DFVT +PPT+ + ILV+VD SK AHF+
Sbjct 1171 CQLIKSHRPRLHGLLQPLPIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFI 1230
Query 101 PIANLTNAS 109
+A+
Sbjct 1231 ATRKTLDAT 1239
> At2g14650
Length=1328
Score = 52.8 bits (125), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/70 (38%), Positives = 42/70 (60%), Gaps = 1/70 (1%)
Query 40 VCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHF 99
VC+ K+ +Q P G+ Q L IP + +++DFV +P + T D+I V+VD L+K AHF
Sbjct 952 VCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRT-KDAIWVIVDRLTKSAHF 1010
Query 100 VPIANLTNAS 109
+ I A+
Sbjct 1011 LAIRKTDGAA 1020
> At3g11970_2
Length=958
Score = 49.7 bits (117), Expect = 2e-06, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 49/94 (52%), Gaps = 5/94 (5%)
Query 19 HIRVHGL--WRICVPQFPEFLTH--VCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVT 74
H RV GL W+ + ++ C+ KS GL Q L IP + VS+DF+
Sbjct 562 HQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIE 621
Query 75 DIPPTTTGHDSILVMVDSLSKMAHFVPIANLTNA 108
+P + G I+V+VD LSK AHF+ +++ +A
Sbjct 622 GLP-VSGGKTVIMVVVDRLSKAAHFIALSHPYSA 654
> At3g31970
Length=1329
Score = 49.3 bits (116), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 39/63 (61%), Gaps = 1/63 (1%)
Query 40 VCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHF 99
VC+ K+ +Q P GL Q L I + +++DFV +P + T D+I V+VD L+K AHF
Sbjct 950 VCQLVKAEHQVPGGLLQSLPILEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTKSAHF 1008
Query 100 VPI 102
+ I
Sbjct 1009 LAI 1011
> At1g36590_2
Length=958
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 29/84 (34%), Positives = 43/84 (51%), Gaps = 12/84 (14%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHFV 100
C+ KS GL Q L IP + VS+DF+ +P + G I+V+VD LSK AHF+
Sbjct 588 CQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLP-VSGGKTVIMVVVDRLSKAAHFI 646
Query 101 PIANLTNASGSVCFRPWNLLTTQQ 124
+++ P++ LT Q
Sbjct 647 ALSH-----------PYSALTVAQ 659
> At4g07830
Length=611
Score = 47.0 bits (110), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query 40 VCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHF 99
VC+ K +Q L Q L IP + +++DFV +P + T D+I V+VD L+K AHF
Sbjct 217 VCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTKSAHF 275
Query 100 VPIANLTNAS 109
+ I A+
Sbjct 276 LAIRKTDGAA 285
> At4g10580
Length=1240
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 24/70 (34%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query 40 VCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAHF 99
VC+ K+ +Q G+ Q L IP + +++D V + + T D+I V+VD L+K AHF
Sbjct 987 VCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGLRVSRT-KDAIWVIVDRLTKSAHF 1045
Query 100 VPIANLTNAS 109
+ I A+
Sbjct 1046 LAIRKTDGAA 1055
> At2g06190
Length=280
Score = 40.8 bits (94), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 70 LDFVTDIPPTTTGHDSILVMVDSLSKMAHFVPIANLTNAS 109
+DFV +P T G DS+ V+VD SKM HF+ +AS
Sbjct 1 MDFVLGLPRTQRGVDSVFVVVDRFSKMTHFIACKKTADAS 40
> At2g06890
Length=1215
Score = 39.7 bits (91), Expect = 0.002, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 34/65 (52%), Gaps = 3/65 (4%)
Query 39 HVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILVMVDSLSKMAH 98
+ C+ +KS Q P GL+ L IP +S+DFV +P TG DSI V+ +
Sbjct 939 NTCKQAKSKIQ-PNGLYTPLPIPKHPWNDISMDFVMGLP--RTGKDSIFVVYSPFQIVYG 995
Query 99 FVPIA 103
F PI+
Sbjct 996 FNPIS 1000
> At4g03840
Length=973
Score = 33.5 bits (75), Expect = 0.12, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDF 72
C+ K+ +Q P+GL Q L IP + H+++DF
Sbjct 658 CQLVKAEHQVPSGLLQNLPIPEWKWDHITMDF 689
> At2g06470
Length=899
Score = 33.5 bits (75), Expect = 0.12, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 41 CRASKSINQKPAGLFQQLLIPSRRGAHVSLDF 72
C+ K+ +Q P+GL Q L IP + H+++DF
Sbjct 658 CQLVKAEHQVPSGLLQNLPIPEWKWDHITMDF 689
> At4g10120
Length=1083
Score = 30.8 bits (68), Expect = 0.69, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 25/50 (50%), Gaps = 8/50 (16%)
Query 8 LLEVPCFSRSLHIRVHGLWRICVPQFPEF----LTHVCRASKSINQKPAG 53
++ +PC SR +I LW P PEF L H+ ++S+ ++ G
Sbjct 312 IIRIPCGSRDKYIPKESLW----PHIPEFVDGALNHIVSIARSLGEQVNG 357
> YGR285c
Length=433
Score = 30.8 bits (68), Expect = 0.83, Method: Composition-based stats.
Identities = 17/62 (27%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 55 FQQLLIPSRRGAHVSLDFVTDIPPTTTGHDSILV-----MVDSLSKMAHFVPIANLTNAS 109
F+ L ++R + S DFV D+PP G D + ++ ++ + PI +L N
Sbjct 152 FETLTDSNKRAQYDSCDFVADVPPPKKGTDYDFYEAWGPVFEAEARFSKKTPIPSLGNKD 211
Query 110 GS 111
S
Sbjct 212 SS 213
> SPBC4.03c
Length=891
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 28/68 (41%), Gaps = 0/68 (0%)
Query 9 LEVPCFSRSLHIRVHGLWRICVPQFPEFLTHVCRASKSINQKPAGLFQQLLIPSRRGAHV 68
LE+P L ++V L QFP H + ++ +F L+ + R
Sbjct 814 LELPTLDTVLSVQVRNLLSSLRMQFPSMALHPQIVRQGLDGSEGEIFSTLVEDNTREGFG 873
Query 69 SLDFVTDI 76
LDF+T I
Sbjct 874 YLDFLTKI 881
> YBL095w
Length=270
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 18/25 (72%), Gaps = 2/25 (8%)
Query 59 LIPSRRG--AHVSLDFVTDIPPTTT 81
L+PS+RG A +SL+F DIP TT
Sbjct 169 LLPSKRGVTARLSLEFFEDIPVDTT 193
> At3g61830
Length=613
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 5/65 (7%)
Query 15 SRSLHIRVH-GLWRICVPQFPEFLTHVCRASKSINQKPAGLFQQLLIPSRRGAHVSLDFV 73
SRS +++ LW++C L V RA + + P G +QL+ + +G + V
Sbjct 15 SRSYQDQLYTELWKVCAGP----LVEVPRAQERVFYFPQGHMEQLVASTNQGINSEEIPV 70
Query 74 TDIPP 78
D+PP
Sbjct 71 FDLPP 75
> Hs17463002
Length=115
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 22/46 (47%), Gaps = 1/46 (2%)
Query 75 DIPPTTT-GHDSILVMVDSLSKMAHFVPIANLTNASGSVCFRPWNL 119
DIPP DS+ + VD+L +A++TNA FR L
Sbjct 16 DIPPEEKLTKDSVTIRVDNLCHQIQNATMADITNADSKPIFRHQEL 61
> CE10950
Length=337
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 20 IRVHGLWRICVPQFPEFLTHVCRASKSINQKPAGLFQQLLIPSRRGAHVSL 70
+ ++ + IC+P FP ++T K INQ Q ++ P + AH L
Sbjct 185 VAIYSIGHICLPFFPVYITIFILRQKIINQLHVK--QHVMSPDTKAAHSQL 233
Lambda K H
0.325 0.135 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1425342594
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40