bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0241_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
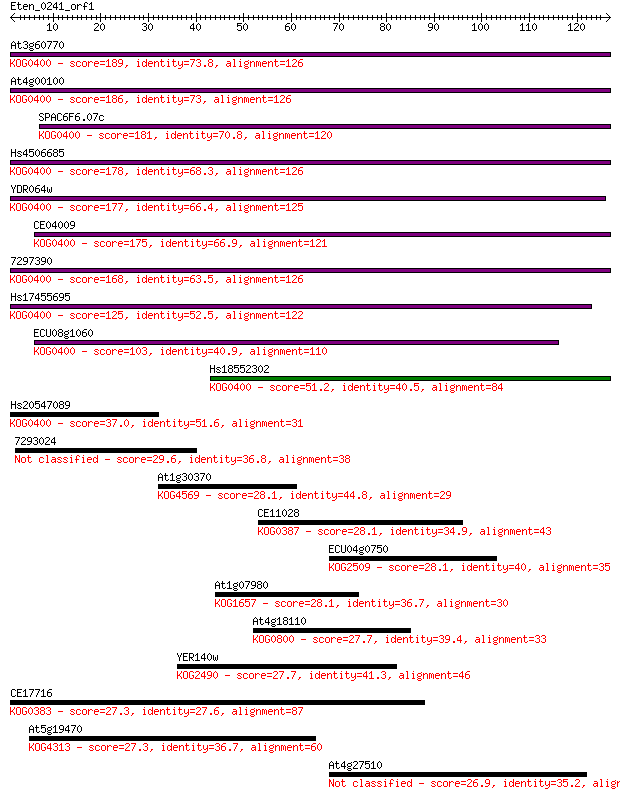
At3g60770 189 1e-48
At4g00100 186 8e-48
SPAC6F6.07c 181 3e-46
Hs4506685 178 2e-45
YDR064w 177 4e-45
CE04009 175 2e-44
7297390 168 2e-42
Hs17455695 125 2e-29
ECU08g1060 103 9e-23
Hs18552302 51.2 5e-07
Hs20547089 37.0 0.009
7293024 29.6 1.5
At1g30370 28.1 3.7
CE11028 28.1 4.1
ECU04g0750 28.1 4.4
At1g07980 28.1 4.5
At4g18110 27.7 5.2
YER140w 27.7 5.9
CE17716 27.3 6.6
At5g19470 27.3 6.9
At4g27510 26.9 8.0
> At3g60770
Length=150
Score = 189 bits (480), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 93/126 (73%), Positives = 107/126 (84%), Gaps = 1/126 (0%)
Query 1 LRVKPQEVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIP 60
L+ PQ+V++ I K A+KG TPSQIGV LRD GIPQVKSVTGSKILRILK G+APEIP
Sbjct 26 LKTTPQDVDESICKFAKKGLTPSQIGVILRDSHGIPQVKSVTGSKILRILKAHGLAPEIP 85
Query 61 EDLYFLIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWKYQSST 120
EDLY LIKKAVA+RKHLE++RKD DSKFRLILVESRIHRL RYY++TK+LP W +S+T
Sbjct 86 EDLYHLIKKAVAIRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPPVWN-ESTT 144
Query 121 ASALVA 126
AS LVA
Sbjct 145 ASTLVA 150
> At4g00100
Length=150
Score = 186 bits (472), Expect = 8e-48, Method: Compositional matrix adjust.
Identities = 92/126 (73%), Positives = 106/126 (84%), Gaps = 1/126 (0%)
Query 1 LRVKPQEVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIP 60
L+ Q+V++ I K A+KG TPSQIGV LRD GIPQVKSVTGSKILRILK G+APEIP
Sbjct 26 LKTTSQDVDESICKFAKKGLTPSQIGVILRDSHGIPQVKSVTGSKILRILKAHGLAPEIP 85
Query 61 EDLYFLIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWKYQSST 120
EDLY LIKKAVA+RKHLE++RKD DSKFRLILVESRIHRL RYY++TK+LP W +S+T
Sbjct 86 EDLYHLIKKAVAIRKHLERNRKDKDSKFRLILVESRIHRLARYYKKTKKLPPVWN-ESTT 144
Query 121 ASALVA 126
AS LVA
Sbjct 145 ASTLVA 150
> SPAC6F6.07c
Length=151
Score = 181 bits (459), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 85/120 (70%), Positives = 102/120 (85%), Gaps = 0/120 (0%)
Query 7 EVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIPEDLYFL 66
V + I+K ++KG +PSQIGVTLRD GIPQV+ +TG KI+RILK G+APE+PEDLY L
Sbjct 32 SVVEQILKFSKKGMSPSQIGVTLRDSHGIPQVRFITGQKIMRILKANGLAPELPEDLYNL 91
Query 67 IKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWKYQSSTASALVA 126
IKKAV+VRKHLE++RKD DSKFRLIL+ESRIHRL RYYR+ LP TWKY+S+TASALVA
Sbjct 92 IKKAVSVRKHLERNRKDKDSKFRLILIESRIHRLARYYRKVGALPPTWKYESATASALVA 151
> Hs4506685
Length=151
Score = 178 bits (452), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 86/126 (68%), Positives = 106/126 (84%), Gaps = 0/126 (0%)
Query 1 LRVKPQEVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIP 60
L++ +V++ I KLA+KG TPSQIGV LRD G+ QV+ VTG+KILRILK +G+AP++P
Sbjct 26 LKLTSDDVKEQIYKLAKKGLTPSQIGVILRDSHGVAQVRFVTGNKILRILKSKGLAPDLP 85
Query 61 EDLYFLIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWKYQSST 120
EDLY LIKKAVAVRKHLE++RKD D+KFRLIL+ESRIHRL RYY+ + LP WKY+SST
Sbjct 86 EDLYHLIKKAVAVRKHLERNRKDKDAKFRLILIESRIHRLARYYKTKRVLPPNWKYESST 145
Query 121 ASALVA 126
ASALVA
Sbjct 146 ASALVA 151
> YDR064w
Length=151
Score = 177 bits (449), Expect = 4e-45, Method: Compositional matrix adjust.
Identities = 83/125 (66%), Positives = 101/125 (80%), Gaps = 0/125 (0%)
Query 1 LRVKPQEVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIP 60
++ + V + IVK ARKG TPSQIGV LRD G+ Q + +TG+KI+RILK G+APEIP
Sbjct 26 FKLSSESVIEQIVKYARKGLTPSQIGVLLRDAHGVTQARVITGNKIMRILKSNGLAPEIP 85
Query 61 EDLYFLIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWKYQSST 120
EDLY+LIKKAV+VRKHLE++RKD D+KFRLIL+ESRIHRL RYYR LP WKY+S+T
Sbjct 86 EDLYYLIKKAVSVRKHLERNRKDKDAKFRLILIESRIHRLARYYRTVAVLPPNWKYESAT 145
Query 121 ASALV 125
ASALV
Sbjct 146 ASALV 150
> CE04009
Length=151
Score = 175 bits (443), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 81/121 (66%), Positives = 102/121 (84%), Gaps = 0/121 (0%)
Query 6 QEVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIPEDLYF 65
+EV+D IVK+A+KG PSQIGV LRD G+ QV+ + G+KI RILK +G+APE+PEDLY
Sbjct 31 EEVQDQIVKMAKKGLRPSQIGVILRDSHGVGQVRRLAGNKIFRILKSKGMAPELPEDLYH 90
Query 66 LIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWKYQSSTASALV 125
L+KKAVA+RKHLE+ RKD DSK+RLILVESRIHRL RYY+ +QLP TWKY+S TA++LV
Sbjct 91 LVKKAVAIRKHLERSRKDIDSKYRLILVESRIHRLARYYKTKRQLPPTWKYESGTAASLV 150
Query 126 A 126
+
Sbjct 151 S 151
> 7297390
Length=151
Score = 168 bits (426), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 80/126 (63%), Positives = 100/126 (79%), Gaps = 0/126 (0%)
Query 1 LRVKPQEVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIP 60
L++ +V++ I KL +KG TPS+IG+ LRD G+ QV+ V G+KILRI+K G+ P+IP
Sbjct 26 LKLNADDVKEQIKKLGKKGLTPSKIGIILRDSHGVAQVRFVNGNKILRIMKSVGLKPDIP 85
Query 61 EDLYFLIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWKYQSST 120
EDLY +IKKAVA+RKHLE++RKD D KFRLILVESRIHRL RYY+ LP WKY+SST
Sbjct 86 EDLYHMIKKAVAIRKHLERNRKDKDGKFRLILVESRIHRLARYYKTKSVLPPNWKYESST 145
Query 121 ASALVA 126
ASALVA
Sbjct 146 ASALVA 151
> Hs17455695
Length=232
Score = 125 bits (314), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 64/122 (52%), Positives = 82/122 (67%), Gaps = 20/122 (16%)
Query 1 LRVKPQEVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIP 60
L++ V++ I KL +KG TP QIG+ RILK +G+AP++P
Sbjct 119 LKLTSDNVKEQIYKLTKKGLTPPQIGI--------------------RILKSKGLAPDLP 158
Query 61 EDLYFLIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWKYQSST 120
EDLY LIKKAVAV+KHLE+ RKD D+KF LIL+ESRIHRL RYY+ + LP +WKY+ ST
Sbjct 159 EDLYHLIKKAVAVQKHLERSRKDKDAKFLLILIESRIHRLARYYKTKRVLPPSWKYEPST 218
Query 121 AS 122
AS
Sbjct 219 AS 220
> ECU08g1060
Length=148
Score = 103 bits (257), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 69/110 (62%), Gaps = 0/110 (0%)
Query 6 QEVEDHIVKLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQGIAPEIPEDLYF 65
E++ ++++ KG IG LRD++GI + V G I R L++ G+ P+IP DL
Sbjct 31 DEIKSDVIQMGNKGVPAPDIGTRLRDEYGIGKASDVLGESITRFLQRNGVVPKIPHDLES 90
Query 66 LIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGRYYRRTKQLPATWK 115
L+ +A +R HL RKD +K+RLILV SR++R+ RYY+R ++P WK
Sbjct 91 LVHRANTLRSHLNIYRKDNSAKYRLILVSSRMYRVARYYKRKMRIPGNWK 140
> Hs18552302
Length=92
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 34/85 (40%), Positives = 42/85 (49%), Gaps = 37/85 (43%)
Query 43 GSKILRILKKQGIAPEIPEDLYFLIKKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGR 102
G+ ILRILK + +AP++PEDLY LIK+A
Sbjct 44 GNTILRILKSKRLAPDLPEDLYCLIKQA-------------------------------- 71
Query 103 YYRRTKQ-LPATWKYQSSTASALVA 126
TKQ LP WK++SSTASALVA
Sbjct 72 ----TKQILPPNWKHESSTASALVA 92
> Hs20547089
Length=97
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 24/31 (77%), Gaps = 0/31 (0%)
Query 1 LRVKPQEVEDHIVKLARKGQTPSQIGVTLRD 31
L++ P ++++ I KLA+KG TPSQ+ V LRD
Sbjct 58 LKLIPDDMKEQIYKLAKKGLTPSQVSVILRD 88
> 7293024
Length=2478
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 25/42 (59%), Gaps = 4/42 (9%)
Query 2 RVKPQEVEDHIVKLARKGQTPSQIGVTLRDQFG----IPQVK 39
R+ P + E + + + PSQ+ VT+RDQ+G +P++K
Sbjct 62 RLDPNKCELNTITEPLRYGWPSQVTVTIRDQYGDAVLVPELK 103
> At1g30370
Length=529
Score = 28.1 bits (61), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 8/37 (21%)
Query 32 QFGIPQVKSVT--------GSKILRILKKQGIAPEIP 60
FG P+V ++ G K+LR++ KQ I P++P
Sbjct 356 SFGAPRVGNLAFKEKLNSLGVKVLRVVNKQDIVPKLP 392
> CE11028
Length=1785
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 25/48 (52%), Gaps = 5/48 (10%)
Query 53 QGIAPEIP-----EDLYFLIKKAVAVRKHLEKDRKDADSKFRLILVES 95
+ + P +P E+L FL+ AV VR H+ K + + K R + V +
Sbjct 1676 EAVHPMVPRTSDREELNFLVAMAVDVRSHMLKCYGETEKKLRELFVRN 1723
> ECU04g0750
Length=429
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 68 KKAVAVRKHLEKDRKDADSKFRLILVESRIHRLGR 102
+ +AV K E DRK + FRL + +RI++L R
Sbjct 25 RDGLAVGKAYELDRKRIEMNFRLDQINTRINQLNR 59
> At1g07980
Length=206
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 44 SKILRILKKQGIAPEIPEDLYFLIKKAVAV 73
++I RI++ AP+I +D FL+ KA +
Sbjct 113 NRIRRIMRSDNSAPQIMQDAVFLVNKATEM 142
> At4g18110
Length=213
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 7/33 (21%)
Query 52 KQGIAPEIPEDLYFLIKKAVAVRKHLEKDRKDA 84
+Q I P+I EDL + +R+HLE++ KD
Sbjct 95 QQSIVPKICEDL-------IDIRRHLEEEEKDG 120
> YER140w
Length=556
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 27/53 (50%), Gaps = 8/53 (15%)
Query 36 PQVKSVTGSKIL-------RILKKQGIAPEIPEDLYFLIKKAVAVRKHLEKDR 81
P V SV GS++L I K I P+I + Y++I K + R H +DR
Sbjct 340 PMV-SVVGSEVLVDWAKHAYITKFNRIRPQIYDKFYYIIYKDYSTRTHKLEDR 391
> CE17716
Length=1829
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 24/94 (25%), Positives = 42/94 (44%), Gaps = 13/94 (13%)
Query 1 LRVKPQEVEDHIV-------KLARKGQTPSQIGVTLRDQFGIPQVKSVTGSKILRILKKQ 53
+VK +E +DH+ K AR +TP+ IP + T ++ R ++
Sbjct 136 FQVKYKEYQDHMAAQGKPVQKQARGSKTPAVSTPV------IPPRSAPTKTRSARRKRRD 189
Query 54 GIAPEIPEDLYFLIKKAVAVRKHLEKDRKDADSK 87
AP+ ++ IK+ + L KD++DA K
Sbjct 190 SDAPDSDQEFEAFIKQQEQLEDDLVKDKEDARIK 223
> At5g19470
Length=373
Score = 27.3 bits (59), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 33/73 (45%), Gaps = 13/73 (17%)
Query 5 PQEVEDHIVKLARKGQTP-----------SQIG--VTLRDQFGIPQVKSVTGSKILRILK 51
P +E+ IV KG T SQ G VTL P+ ++ + +++IL
Sbjct 98 PFVIEEQIVGYIHKGFTKYLRDFNDIFTFSQYGGHVTLNMMLDKPEERTRAVAHVIKILG 157
Query 52 KQGIAPEIPEDLY 64
+GI P I +LY
Sbjct 158 NKGIIPGIRNELY 170
> At4g27510
Length=677
Score = 26.9 bits (58), Expect = 8.0, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 28/63 (44%), Gaps = 9/63 (14%)
Query 68 KKAVAVRKHLEKDRKDADSKFRLILVESRI---HR-----LGRYYRRTKQLPATW-KYQS 118
K V ++H+ K KF + +SR+ HR L + T+QL +W YQS
Sbjct 326 KSVVLYKRHMSKSENGLSGKFGFLTSKSRVAFEHRLLPSTLAKVNDVTQQLKDSWDSYQS 385
Query 119 STA 121
A
Sbjct 386 DNA 388
Lambda K H
0.320 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40