bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0252_orf2
Length=215
Score E
Sequences producing significant alignments: (Bits) Value
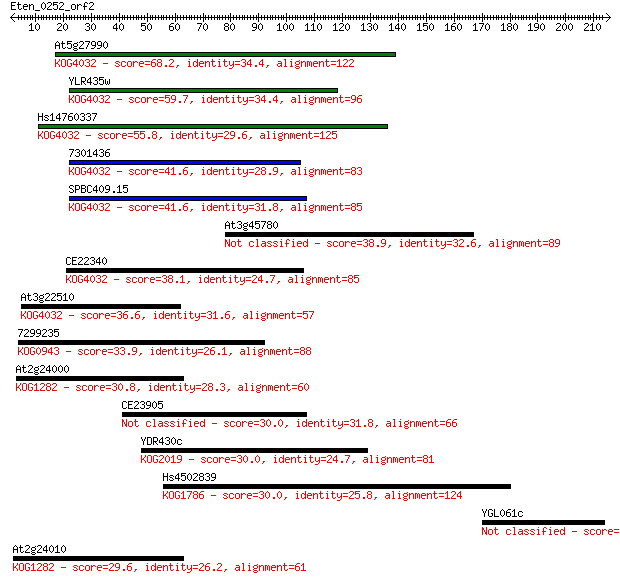
At5g27990 68.2 1e-11
YLR435w 59.7 4e-09
Hs14760337 55.8 6e-08
7301436 41.6 0.001
SPBC409.15 41.6 0.001
At3g45780 38.9 0.009
CE22340 38.1 0.014
At3g22510 36.6 0.040
7299235 33.9 0.23
At2g24000 30.8 1.9
CE23905 30.0 3.4
YDR430c 30.0 3.4
Hs4502839 30.0 3.8
YGL061c 30.0 4.0
At2g24010 29.6 4.4
> At5g27990
Length=184
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 42/124 (33%), Positives = 65/124 (52%), Gaps = 3/124 (2%)
Query 17 EAEPIFREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLIAEFRENKN--VDVED 74
EA I +E + +L +WT + AVE GWGGRDS K + + F ++K+ D+E
Sbjct 10 EARDILKEGIGLILWRWTAMRAAVENGWGGRDSQAKANETVATVFDFFIQSKDPVKDIEK 69
Query 75 LACTLSERLASDFSVSVEDDSDLEVAQLLVDLHEQISRGCFDLASVVKQQQNQRATSSAK 134
L L + L + + + ED S EVA LLVDL+E G +++ ++ +Q + S K
Sbjct 70 LGDLLDKGL-DELNTTAEDGSVDEVANLLVDLYEDCCNGNYEMLEELRATYSQTSASVVK 128
Query 135 SQCG 138
G
Sbjct 129 VSNG 132
> YLR435w
Length=249
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 33/96 (34%), Positives = 52/96 (54%), Gaps = 4/96 (4%)
Query 22 FREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLIAEFRENKNVDVEDLACTLSE 81
F VS V+ +W L++AVE WGG DS KR + ++ F+ K VD + TL
Sbjct 76 FELGVSMVIYKWDALDVAVENSWGGPDSAEKRDWITGIVVDLFKNEKVVDAALIEETLLY 135
Query 82 RLASDFSVSVEDDSDLEVAQLLVDLHEQISRGCFDL 117
+ +F +VEDDS L +A +++++ CF+L
Sbjct 136 AMIDEFETNVEDDSALPIAVEVINIY----NDCFNL 167
> Hs14760337
Length=191
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 65/129 (50%), Gaps = 4/129 (3%)
Query 11 MHQRRQEAEPIFREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLIAEFRENKNV 70
M ++A +FR V A L W L +AVE G+GG S K + L + F N ++
Sbjct 1 MAGAAEDARALFRAGVCAALEAWPALQIAVENGFGGVHSQEKAKWLGGAVEDYFMRNADL 60
Query 71 DVEDLACTLSERLASDFSVSVEDDSDLEVAQLLVDLHEQISRG----CFDLASVVKQQQN 126
+++++ L E L ++F VED S +V+Q L + RG ++AS + Q++
Sbjct 61 ELDEVEDFLGELLTNEFDTVVEDGSLPQVSQQLQTMFHHFQRGDGAALREMASCITQRKC 120
Query 127 QRATSSAKS 135
+ ++ K+
Sbjct 121 KVTATALKT 129
> 7301436
Length=195
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 24/83 (28%), Positives = 40/83 (48%), Gaps = 0/83 (0%)
Query 22 FREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLIAEFRENKNVDVEDLACTLSE 81
FR V + W L LAVE G GGR+ + ++ + N+N+ +L L E
Sbjct 13 FRVIVEKIFNHWQDLRLAVEHGMGGRNGQQVAIEIMDYTYQYCVSNENITQGELVEVLEE 72
Query 82 RLASDFSVSVEDDSDLEVAQLLV 104
+ +F+ +DDS E+ + L+
Sbjct 73 LMDQEFNTLCDDDSIPEICRNLL 95
> SPBC409.15
Length=179
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 45/86 (52%), Gaps = 1/86 (1%)
Query 22 FREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLIAEFRENKNVDVEDLACTLSE 81
F AV +L W ++ AVE+ W D+ KR + L+ +V+ D+ + +
Sbjct 12 FEYAVGVLLCSWPVMKQAVEEEWADVDTADKRDWMAGVLVDYITVTSDVEAWDVEELILQ 71
Query 82 RLASDFSV-SVEDDSDLEVAQLLVDL 106
L +F+V S+EDDS +AQ LV++
Sbjct 72 VLQDEFNVGSIEDDSPYILAQDLVNV 97
> At3g45780
Length=996
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 44/96 (45%), Gaps = 7/96 (7%)
Query 78 TLSERLASDFSVSVEDDSDLEVAQLLVDLHEQISR---GCFDLASVVKQQQNQRATSSAK 134
T + L D + L+V QL+ L ++ + GCF+ A+ VKQ + + A
Sbjct 900 TFTNVLQKDLKFPASIPASLQVKQLIFRLLQRDPKKRLGCFEGANEVKQHSFFKGINWAL 959
Query 135 SQCGNAREAETDCSMSEGSDAE----GELEDLISSV 166
+C N E ET E + E ELEDL ++V
Sbjct 960 IRCTNPPELETPIFSGEAENGEKVVDPELEDLQTNV 995
> CE22340
Length=225
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 21/85 (24%), Positives = 40/85 (47%), Gaps = 0/85 (0%)
Query 21 IFREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLIAEFRENKNVDVEDLACTLS 80
+ R L W+ LA++ GG ++ K + E L + + + E+L L+
Sbjct 58 VLRNFTGRTLKSWSGYQLALDNSCGGDETREKDKWFLEVLCEQLTTTRGLKAEELEEWLT 117
Query 81 ERLASDFSVSVEDDSDLEVAQLLVD 105
L DF + ++DDS ++A L++
Sbjct 118 NILYHDFDLILDDDSSYQIAFFLLE 142
> At3g22510
Length=85
Score = 36.6 bits (83), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Query 5 PQQHQKMHQRRQEAEPIFREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLI 61
P H K H + + + + + +L++W L +AV+ WGG DS +K Q+L +L
Sbjct 15 PTWHHK-HVPPMDDDSVLQRDIGELLSRWGGLQMAVKNQWGGHDSLKKSQELAHNLF 70
> 7299235
Length=2165
Score = 33.9 bits (76), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 23/88 (26%), Positives = 40/88 (45%), Gaps = 1/88 (1%)
Query 4 RPQQHQKMHQRRQEAEPIFREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLIAE 63
R Q+ + R +EAE + R V +V + + NL + R SH K + +
Sbjct 1459 RELQNASVSNRSKEAEEVVRRFVRSVARVFVIFNLEKQPNPEKRKSHSSCNKYVQSCVKV 1518
Query 64 FRENKNVDVEDLACTLSERLASDFSVSV 91
F+ + +E+L C +SE L + + V
Sbjct 1519 FQTLHKISIEEL-CEVSEALIAPVRLGV 1545
> At2g24000
Length=474
Score = 30.8 bits (68), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 27/60 (45%), Gaps = 11/60 (18%)
Query 3 NRPQQHQKMHQRRQEAEPIFREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLIA 62
NRP+ + MH +A+ +WT + +V W RDS +Y++LIA
Sbjct 334 NRPEVQRAMHANH-----------TAIPYKWTACSDSVFNNWNWRDSDNSMLPIYKELIA 382
> CE23905
Length=301
Score = 30.0 bits (66), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 32/68 (47%), Gaps = 7/68 (10%)
Query 41 EQGWGGRDSHRKRQKLY--EDLIAEFRENKNVDVEDLACTLSERLASDFSVSVEDDSDLE 98
E G+ +RK KL +D EFRE V +L L FS++ E D D++
Sbjct 73 EDGYFLHRRNRKLTKLIKTQDFYTEFRE-----VFNLLLKYQRALLQRFSINFESDGDIK 127
Query 99 VAQLLVDL 106
+ + LV +
Sbjct 128 IFEELVHI 135
> YDR430c
Length=989
Score = 30.0 bits (66), Expect = 3.4, Method: Composition-based stats.
Identities = 20/82 (24%), Positives = 42/82 (51%), Gaps = 8/82 (9%)
Query 48 DSHRKRQKLYEDLIAEFRENKNVDVEDLAC-TLSERLASDFSVSVEDDSDLEVAQLLVDL 106
D K L++DLI ++ +K C T S + + +FS S++D+ + + + L
Sbjct 452 DLETKGDTLFQDLIRKYIVHK-------PCFTFSIQGSEEFSKSLDDEEQTRLREKITAL 504
Query 107 HEQISRGCFDLASVVKQQQNQR 128
EQ + F +++++QN++
Sbjct 505 DEQDKKNIFKRGILLQEKQNEK 526
> Hs4502839
Length=3801
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 60/128 (46%), Gaps = 11/128 (8%)
Query 56 LYEDLIAEFREN-KNVDVEDLACTLSERLASDFSVSVEDDSDLEVAQ-LLVDLHEQI--S 111
++E + R+ KNV + T+ L S+ +DDSD + Q +LVDL + S
Sbjct 1292 VFESFLKIIRQKEKNVFLLMQQGTVKNLLGGFLSILTQDDSDFQACQRVLVDLLVSLMSS 1351
Query 112 RGCFDLASVVKQQQNQRATSSAKSQCGNAREAETDCSMSEGSDAEGELEDLISSVEIAPS 171
R C + +++ + +++ + G + E+D +MS + L + AP+
Sbjct 1352 RTCSEELTLLLRIFLEKSPCTKILLLGILKIIESDTTMSPS-------QYLTFPLLHAPN 1404
Query 172 PSEGIASQ 179
S G++SQ
Sbjct 1405 LSNGVSSQ 1412
> YGL061c
Length=247
Score = 30.0 bits (66), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 22/44 (50%), Gaps = 0/44 (0%)
Query 170 PSPSEGIASQTRSRQTAGAASELQPAHPSEADGWTTVQAGSRRR 213
P PS GI++ R R+T AS+ P + WT A S R+
Sbjct 201 PVPSAGISNNGRVRKTHVPASKRPSGIPRVTNRWTKPTASSSRK 244
> At2g24010
Length=425
Score = 29.6 bits (65), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 27/61 (44%), Gaps = 13/61 (21%)
Query 2 QNRPQQHQKMHQRRQEAEPIFREAVSAVLAQWTLLNLAVEQGWGGRDSHRKRQKLYEDLI 61
NRP + MH ++++ +WTL N+ V W +DS +Y++L
Sbjct 286 YNRPDVQRAMHAN-----------LTSIPYKWTLCNMVVNNNW--KDSEFSMLPIYKELT 332
Query 62 A 62
A
Sbjct 333 A 333
Lambda K H
0.310 0.124 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3969005466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40