bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
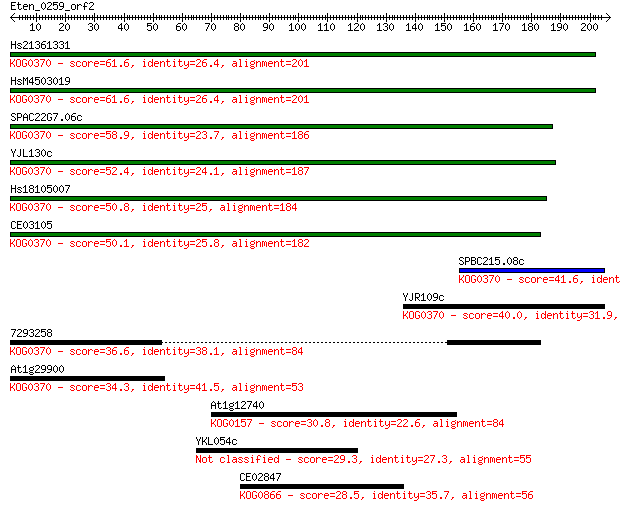
Query= Eten_0259_orf2
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
Hs21361331 61.6 9e-10
HsM4503019 61.6 1e-09
SPAC22G7.06c 58.9 6e-09
YJL130c 52.4 6e-07
Hs18105007 50.8 2e-06
CE03105 50.1 3e-06
SPBC215.08c 41.6 0.001
YJR109c 40.0 0.003
7293258 36.6 0.038
At1g29900 34.3 0.16
At1g12740 30.8 2.1
YKL054c 29.3 5.5
CE02847 28.5 8.7
> Hs21361331
Length=1500
Score = 61.6 bits (148), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 53/201 (26%), Positives = 88/201 (43%), Gaps = 55/201 (27%)
Query 1 AMVSAGLRLPLMRRSLMLVTGPRWSKVDFFPFALKLVKLGFTIYATEGTYNFLRKSLRGC 60
AM+S G ++P + +L+ + + F A +L GF ++ATE T ++L
Sbjct 1349 AMLSTGFKIP---QKGILIGIQQSFRPRFLGVAEQLHNEGFKLFATEATSDWLNA----- 1400
Query 61 LEAVSEETGSAQRNTEPWSSLAAPVESGEELETPPGEMVLSEIEKQIISVSKDGNSAEAG 120
N P + +A P + G+ LS I K
Sbjct 1401 -------------NNVPATPVAWPSQEGQNPS-------LSSIRK--------------- 1425
Query 121 GQDLLTATDVIKRGLVELVINVPGRAHHRSVSAGYIIRRTAVDAGVPLLTDLKLASFFVE 180
+I+ G ++LVIN+P + + V Y+IRRTAVD+G+PLLT+ ++ F E
Sbjct 1426 ---------LIRDGSIDLVINLPNN-NTKFVHDNYVIRRTAVDSGIPLLTNFQVTKLFAE 1475
Query 181 SLSKKISRERGRKRFWEVRSW 201
++ K SR+ K + R +
Sbjct 1476 AVQK--SRKVDSKSLFHYRQY 1494
> HsM4503019
Length=1500
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 53/201 (26%), Positives = 88/201 (43%), Gaps = 55/201 (27%)
Query 1 AMVSAGLRLPLMRRSLMLVTGPRWSKVDFFPFALKLVKLGFTIYATEGTYNFLRKSLRGC 60
AM+S G ++P + +L+ + + F A +L GF ++ATE T ++L
Sbjct 1349 AMLSTGFKIP---QKGILIGIQQSFRPRFLGVAEQLHNEGFKLFATEATSDWLNA----- 1400
Query 61 LEAVSEETGSAQRNTEPWSSLAAPVESGEELETPPGEMVLSEIEKQIISVSKDGNSAEAG 120
N P + +A P + G+ LS I K
Sbjct 1401 -------------NNVPANPVAWPSQEGQNPS-------LSSIRK--------------- 1425
Query 121 GQDLLTATDVIKRGLVELVINVPGRAHHRSVSAGYIIRRTAVDAGVPLLTDLKLASFFVE 180
+I+ G ++LVIN+P + + V Y+IRRTAVD+G+PLLT+ ++ F E
Sbjct 1426 ---------LIRDGSIDLVINLPNN-NTKFVHDNYVIRRTAVDSGIPLLTNFQVTKLFAE 1475
Query 181 SLSKKISRERGRKRFWEVRSW 201
++ K SR+ K + R +
Sbjct 1476 AVQK--SRKVDSKSLFHYRQY 1494
> SPAC22G7.06c
Length=2244
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 44/190 (23%), Positives = 80/190 (42%), Gaps = 56/190 (29%)
Query 1 AMVSAGLRLPLMRRSLMLVTGPRWSKVDFFPFALKLVKLGFTIYATEGTYNFLRKSLRGC 60
AM+S G RLP ++++++ G K + P+ KL + + I+AT GT ++ +S C
Sbjct 1384 AMISTGFRLP--KKNILISIGSYKEKAELLPYVKKLYENNYNIFATAGTSDYFMESGVPC 1441
Query 61 LEAVSEETGSAQRNTEPWSSLAAPVESGEELETPPGEMVLSEIEKQIISVSKDGNSAEAG 120
+ L P E EA
Sbjct 1442 ----------------------------KYLADLPAE--------------------EAN 1453
Query 121 GQDLLTATDVIKRGLVELVINVPGRAHHR----SVSAGYIIRRTAVDAGVPLLTDLKLAS 176
+ L+A + ++++ IN+P +R +S+GY RR A+D VPL+T++K A
Sbjct 1454 NEYSLSAH--LANNMIDMYINLPSNNRYRRPANYISSGYKSRRLAIDYSVPLVTNVKCAK 1511
Query 177 FFVESLSKKI 186
+E++ + +
Sbjct 1512 LMIEAICRNL 1521
> YJL130c
Length=2214
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 45/191 (23%), Positives = 79/191 (41%), Gaps = 54/191 (28%)
Query 1 AMVSAGLRLPLMRRSLMLVTGPRWSKVDFFPFALKLVKLGFTIYATEGTYNFLRKSLRGC 60
++++ G +LP +++++L G K + KL +G+ ++AT GT +FL
Sbjct 1350 SLLATGFKLP--KKNILLSIGSYKEKQELLSSVQKLYNMGYKLFATSGTADFL------- 1400
Query 61 LEAVSEETGSAQRNTEPWSSLAAPVESGEELETPPGEMVLSEIEKQIISVSKDGNSAEAG 120
E G A + E +++ D +E
Sbjct 1401 -----SEHGIAVQYLE------------------------------VLNKDDDDQKSE-- 1423
Query 121 GQDLLTATDVIKRGLVELVINVPGRAHHRS----VSAGYIIRRTAVDAGVPLLTDLKLAS 176
+ T + ++L IN+P R VS GY RR AVD VPL+T++K A
Sbjct 1424 ----YSLTQHLANNEIDLYINLPSANRFRRPASYVSKGYKTRRLAVDYSVPLVTNVKCAK 1479
Query 177 FFVESLSKKIS 187
+E++S+ I+
Sbjct 1480 LLIEAISRNIT 1490
> Hs18105007
Length=2225
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 46/189 (24%), Positives = 75/189 (39%), Gaps = 53/189 (28%)
Query 1 AMVSAGLRLPLMRRSLMLVTGPRWSKVDFFPFALKLVKLGFTIYATEGTYNFLRKSLRGC 60
AM+S G ++P +++++L G +K + P L LG+++YA+ GT +F +
Sbjct 1302 AMLSTGFKIP--KKNILLTIGSYKNKSELLPTVRLLESLGYSLYASLGTADFYTEH---- 1355
Query 61 LEAVSEETGSAQRNTEPWSSLAAPVESGEELETPPGEMVLSEIEKQIISVSKDGNSAEAG 120
+ W E + E PP +L ++ ++
Sbjct 1356 ---------GVKVTAVDWH-----FEEAVDGECPPQRSILEQLAEKNF------------ 1389
Query 121 GQDLLTATDVIKRGLVELVINVP-----GRAHHRSVSAGYIIRRTAVDAGVPLLTDLKLA 175
ELVIN+ GR V+ GY RR A D VPL+ D+K
Sbjct 1390 ----------------ELVINLSMRGAGGRRLSSFVTKGYRTRRLAADFSVPLIIDIKCT 1433
Query 176 SFFVESLSK 184
FVE+L +
Sbjct 1434 KLFVEALGQ 1442
> CE03105
Length=2198
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 47/190 (24%), Positives = 82/190 (43%), Gaps = 57/190 (30%)
Query 1 AMVSAGLRLPLMRRSLMLVTGPRWSKVDFFPFALKLVKLGFTIYATEGTYNFLRKSLRGC 60
A++S G +P ++++ + G +K + L+KLG+ +Y ++GT ++ +
Sbjct 1312 ALLSTGFVVP--KQNIFISIGGYHAKAEMLKSVEALLKLGYELYGSKGTADYFQ------ 1363
Query 61 LEAVSEETGSAQRNTEP--WSSLAAPVESGEELETPPGEMVLSEIEKQIISVSKDGNSAE 118
S + N +P W P E +G+S E
Sbjct 1364 ---------SNKINVKPVDW-----PFE--------------------------EGSSDE 1383
Query 119 AGGQDLLTATDVIKRGLVELVINVP--GRAHHRSVSA----GYIIRRTAVDAGVPLLTDL 172
+ + ++ LVIN+P G +R VSA GY RR A+D G+PL+TD+
Sbjct 1384 KTASGTRSVVEFLENKEFHLVINLPIRGSGAYR-VSAFRTHGYKTRRMAIDNGIPLITDI 1442
Query 173 KLASFFVESL 182
K A F+++L
Sbjct 1443 KCAKTFIQAL 1452
> SPBC215.08c
Length=1160
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/52 (40%), Positives = 30/52 (57%), Gaps = 2/52 (3%)
Query 155 YIIRRTAVDAGVPLLTDLKLASFFVESLSKKISRERGRKRFW--EVRSWDEY 204
Y++RR AVD V L+ D+ A FVESL +K+ K+ EV+ W E+
Sbjct 1103 YVMRRNAVDFNVTLINDVNCAKLFVESLKEKLPSVLSEKKEMPSEVKRWSEW 1154
> YJR109c
Length=1118
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 36/75 (48%), Gaps = 6/75 (8%)
Query 136 VELVINVPGRAHHRSVSAGYIIRRTAVDAGVPLLTDLKLASFFVESLSKKISRERGRKRF 195
++ V N+ + + YI+RR A+D +PL + + A F + L KI+ +
Sbjct 1038 IKAVFNLASKRAESTDDVDYIMRRNAIDFAIPLFNEPQTALLFAKCLKAKIAEKIKILES 1097
Query 196 W------EVRSWDEY 204
EVRSWDE+
Sbjct 1098 HDVIVPPEVRSWDEF 1112
> 7293258
Length=2189
Score = 36.6 bits (83), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 31/52 (59%), Gaps = 2/52 (3%)
Query 1 AMVSAGLRLPLMRRSLMLVTGPRWSKVDFFPFALKLVKLGFTIYATEGTYNF 52
AM+S G ++P + +++L G K++ P L K+G+ +YA+ GT +F
Sbjct 1316 AMMSTGFQIP--KNAVLLSIGSFKHKMELLPSIRDLAKMGYKLYASMGTGDF 1365
Score = 35.8 bits (81), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 151 VSAGYIIRRTAVDAGVPLLTDLKLASFFVESL 182
++ GY RR AVD +PL+TD+K VES+
Sbjct 1389 MTHGYRTRRLAVDYSIPLVTDVKCTKLLVESM 1420
> At1g29900
Length=956
Score = 34.3 bits (77), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 32/56 (57%), Gaps = 8/56 (14%)
Query 1 AMVSAGLRLPL---MRRSLMLVTGPRWSKVDFFPFALKLVKLGFTIYATEGTYNFL 53
A ++AG +LPL + SL +T P K+ A+ ++LGF I AT GT +FL
Sbjct 805 AQIAAGQKLPLSGTVFLSLNDMTKPHLEKI-----AVSFLELGFKIVATSGTAHFL 855
> At1g12740
Length=478
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 19/84 (22%), Positives = 37/84 (44%), Gaps = 10/84 (11%)
Query 70 SAQRNTEPWSSLAAPVESGEELETPPGEMVLSEIEKQIISVSKDGNSAEAGGQDLLTATD 129
+A + E WS+ + EL+ M+ K++IS D +S ++L
Sbjct 151 TANKRLELWSN-----QDSVELKDATASMIFDLTAKKLISHDPDKSS-----ENLRANFV 200
Query 130 VIKRGLVELVINVPGRAHHRSVSA 153
+GL+ ++PG A+H+ +
Sbjct 201 AFIQGLISFPFDIPGTAYHKCLQG 224
> YKL054c
Length=738
Score = 29.3 bits (64), Expect = 5.5, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query 65 SEETGSAQRNTEPWSSLAAPVESGEELETPPGEMVLSEIEKQIISVSKDGNSAEA 119
+ T ++Q W+++A P + P E V +E++K+I + KD +EA
Sbjct 231 TSRTSASQPKKMSWAAIATPKPKAVKKTESPLENV-AELKKEISDIKKDDQKSEA 284
> CE02847
Length=340
Score = 28.5 bits (62), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 35/59 (59%), Gaps = 5/59 (8%)
Query 80 SLAAPVESGEELETPPGEMVL-SEIEKQIISVSKDGNSAEAGGQD--LLTATDVIKRGL 135
S APV + +E+E P V S+I ++ V+K+G+S + GG D +TAT++ + +
Sbjct 51 SADAPVANEKEVEEAPQAFVFGSKISDRV--VNKEGDSTKPGGSDEKTMTATELFQNAV 107
Lambda K H
0.317 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3694581778
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40