bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0262_orf1
Length=238
Score E
Sequences producing significant alignments: (Bits) Value
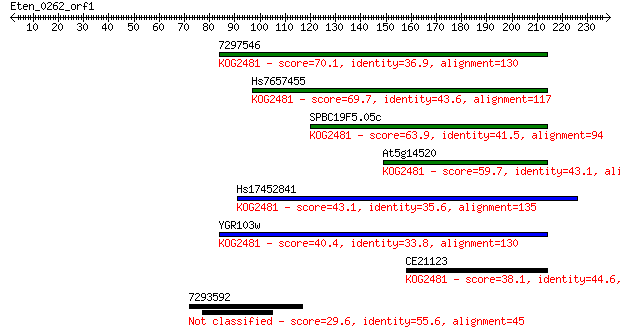
7297546 70.1 4e-12
Hs7657455 69.7 4e-12
SPBC19F5.05c 63.9 3e-10
At5g14520 59.7 5e-09
Hs17452841 43.1 5e-04
YGR103w 40.4 0.004
CE21123 38.1 0.014
7293592 29.6 5.3
> 7297546
Length=627
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 48/133 (36%), Positives = 63/133 (47%), Gaps = 16/133 (12%)
Query 84 SAAEDDSAAEDESAAEDESAAEDESAAEDESAADSSSSEDEDTEEEAEEEAAEDKAEEAA 143
S ++ S D AA + + EDE E+ D + E E+ + K
Sbjct 261 SLKDEASFVSDRIAALNFELLRTDKVQEDE--------EELDIDMELLEQDGDSKRIIKM 312
Query 144 KETESEVEICEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSE---GSPFEL 200
K+ EV ++ LFKGL FF+ RE P PL IRS GG V W S GS ++
Sbjct 313 KQEAQEVS-----RLRTLFKGLKFFINREVPREPLVILIRSFGGKVSWDSSIFAGSTYDE 367
Query 201 GDNRITHQVVDRP 213
GD ITHQ+VDRP
Sbjct 368 GDETITHQIVDRP 380
> Hs7657455
Length=588
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 51/124 (41%), Positives = 67/124 (54%), Gaps = 9/124 (7%)
Query 97 AAEDESAAEDESAAEDESAADSSSSED--EDTEEEAEEEAAEDKAEEAAKE--TESEVEI 152
A E A + ES E +A +S + TEEEAE + E +A+E E+E
Sbjct 260 AGEGTYALDSESCMEKLAALSASLARVVVPATEEEAEVDEFPTDGEMSAQEEDRRKELEA 319
Query 153 CEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSE---GSPFELGDNRITHQV 209
E H + LF+GL FFL RE P L F IRS GG V W G+ +++ D+RITHQ+
Sbjct 320 QEKH--KKLFEGLKFFLNREVPREALAFIIRSFGGEVSWDKSLCIGATYDVTDSRITHQI 377
Query 210 VDRP 213
VDRP
Sbjct 378 VDRP 381
> SPBC19F5.05c
Length=607
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 47/98 (47%), Gaps = 4/98 (4%)
Query 120 SSEDEDTEEEAEEEAAEDKAEEAAKETESEVEICEPHPVQN-LFKGLVFFLCRETPLLPL 178
++ED D E E EE E K + A +S+ + LF FFL RE P L
Sbjct 310 TTEDADEEPETEENLDEFKPADGADNEDSKSLVSHISSSNTSLFSNFTFFLSREVPRFSL 369
Query 179 CFAIRSCGGAVGW---QSEGSPFELGDNRITHQVVDRP 213
F IR+ GG VGW GSPF D ITH + DRP
Sbjct 370 EFVIRAFGGKVGWDPILGSGSPFSESDPVITHHICDRP 407
> At5g14520
Length=590
Score = 59.7 bits (143), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 38/65 (58%), Gaps = 5/65 (7%)
Query 149 EVEICEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSEGSPFELGDNRITHQ 208
E +C ++LFK L FFL RE P L I + GG V W+ EG+PF+ D ITH
Sbjct 329 ETRVC-----KSLFKDLKFFLSREVPRESLQLVITAFGGMVSWEGEGAPFKEDDESITHH 383
Query 209 VVDRP 213
++D+P
Sbjct 384 IIDKP 388
> Hs17452841
Length=505
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 48/155 (30%), Positives = 73/155 (47%), Gaps = 23/155 (14%)
Query 91 AAEDESAAEDESAAEDESAAEDESAADSSSSED--EDTEEEAEEEAAEDKAEEAAKETE- 147
A E+ A+E A + ES+ E +A +S + TEEE E + E +A+E E
Sbjct 205 AQEEAKASEGTHALDSESSMEKMAALTASLARVVVPATEEEPEVDEFPANGEMSAQEEER 264
Query 148 -SEVEICEPHPVQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSE---GSPFELGDN 203
E+E E H + +GL FFL + P L F I S G V W G+ +++ D+
Sbjct 265 RKELEGQEKH--KKFSEGLKFFLNQGVPREALAFVI-SFWGEVSWDKSLCIGATYDITDS 321
Query 204 RITHQVVDRP-------------PEYVLQQCGADL 225
RITH++V+ P P++V+ A L
Sbjct 322 RITHKIVNWPGQQTSIIGRCYMQPQWVIDSVNARL 356
> YGR103w
Length=605
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 44/150 (29%), Positives = 76/150 (50%), Gaps = 20/150 (13%)
Query 84 SAAEDDSAAEDESAAEDESAAEDESAAEDESAADSSSSEDEDTEEEAEEEAAEDKAEEAA 143
S ED D + E++ E A+ +SA ++ + ++TE+E E+E ++K +E
Sbjct 266 SRQEDSLLKLDPTEIEEDVKVESLDASTLKSALNADEANTDETEKEEEQEKKQEKEQEKE 325
Query 144 KETESEVE------------ICEP----HPVQNLFKGLVFFLCRETPLLPLCFAIRSCGG 187
+ E+E++ + +P PV +LF VF++ RE P+ L F I SCGG
Sbjct 326 QNEETELDTFEDNNKNKGDILIQPSKYDSPVASLFSAFVFYVSREVPIDILEFLILSCGG 385
Query 188 AVGWQSEGSPFE----LGDNRITHQVVDRP 213
V ++ E + +++THQ+VDRP
Sbjct 386 NVISEAAMDQIENKKDIDMSKVTHQIVDRP 415
> CE21123
Length=531
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 25/56 (44%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 158 VQNLFKGLVFFLCRETPLLPLCFAIRSCGGAVGWQSEGSPFELGDNRITHQVVDRP 213
++ +FKG VF+L RE P L F IR+ GG VGW+ + + I+H VVDRP
Sbjct 310 IKTMFKGCVFYLNRECPKEALTFIIRNGGGIVGWEGGPTDLKADSKNISHHVVDRP 365
> 7293592
Length=2037
Score = 29.6 bits (65), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 16/28 (57%), Positives = 22/28 (78%), Gaps = 0/28 (0%)
Query 77 EAAAEDDSAAEDDSAAEDESAAEDESAA 104
+ A EDD AAE+D AAE++ AAE++ AA
Sbjct 1279 QGAEEDDQAAENDQAAENDQAAENDQAA 1306
Score = 29.6 bits (65), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 29/45 (64%), Gaps = 0/45 (0%)
Query 72 DDDDDEAAAEDDSAAEDDSAAEDESAAEDESAAEDESAAEDESAA 116
D+D D DD+ + A ED+ AAE++ AAE++ AAE++ AA
Sbjct 1262 DEDRDVKNMIDDAEPMEQGAEEDDQAAENDQAAENDQAAENDQAA 1306
Lambda K H
0.296 0.115 0.298
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4740636838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40