bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0269_orf1
Length=234
Score E
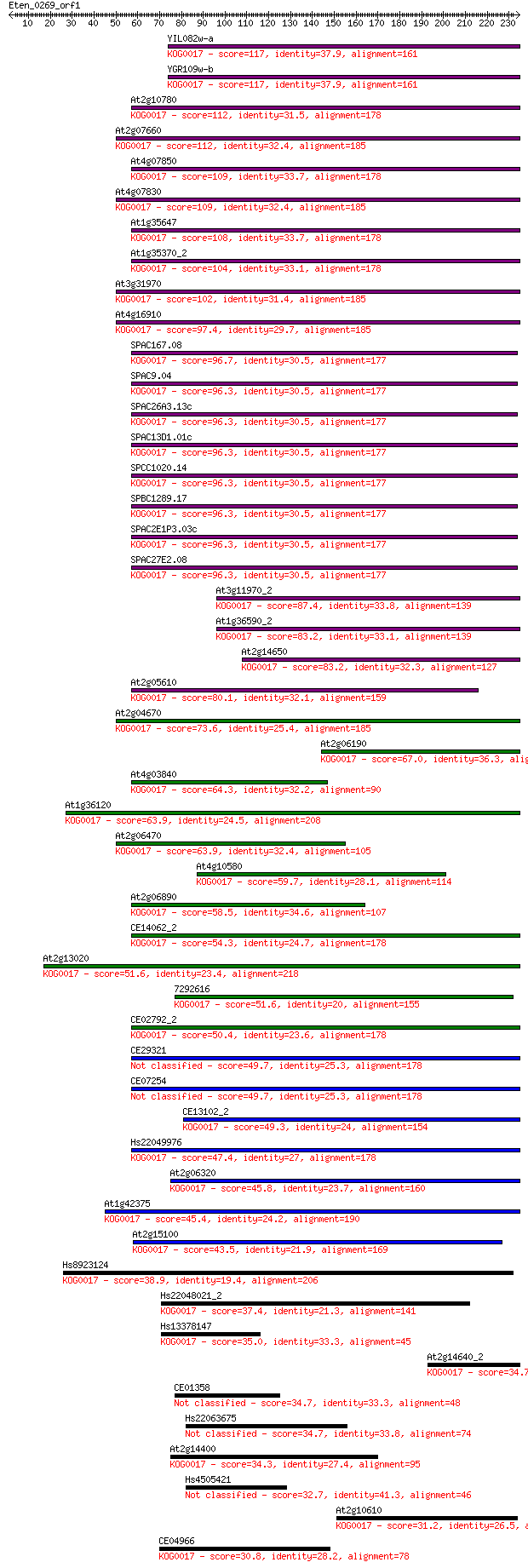
Sequences producing significant alignments: (Bits) Value
YIL082w-a 117 1e-26
YGR109w-b 117 2e-26
At2g10780 112 5e-25
At2g07660 112 1e-24
At4g07850 109 5e-24
At4g07830 109 6e-24
At1g35647 108 1e-23
At1g35370_2 104 1e-22
At3g31970 102 5e-22
At4g16910 97.4 2e-20
SPAC167.08 96.7 4e-20
SPAC9.04 96.3 5e-20
SPAC26A3.13c 96.3 5e-20
SPAC13D1.01c 96.3 5e-20
SPCC1020.14 96.3 5e-20
SPBC1289.17 96.3 5e-20
SPAC2E1P3.03c 96.3 5e-20
SPAC27E2.08 96.3 5e-20
At3g11970_2 87.4 2e-17
At1g36590_2 83.2 5e-16
At2g14650 83.2 5e-16
At2g05610 80.1 3e-15
At2g04670 73.6 4e-13
At2g06190 67.0 3e-11
At4g03840 64.3 2e-10
At1g36120 63.9 3e-10
At2g06470 63.9 3e-10
At4g10580 59.7 5e-09
At2g06890 58.5 1e-08
CE14062_2 54.3 2e-07
At2g13020 51.6 1e-06
7292616 51.6 2e-06
CE02792_2 50.4 3e-06
CE29321 49.7 6e-06
CE07254 49.7 6e-06
CE13102_2 49.3 7e-06
Hs22049976 47.4 2e-05
At2g06320 45.8 8e-05
At1g42375 45.4 1e-04
At2g15100 43.5 3e-04
Hs8923124 38.9 0.009
Hs22048021_2 37.4 0.023
Hs13378147 35.0 0.14
At2g14640_2 34.7 0.16
CE01358 34.7 0.17
Hs22063675 34.7 0.18
At2g14400 34.3 0.21
Hs4505421 32.7 0.59
At2g10610 31.2 1.7
CE04966 30.8 2.2
> YIL082w-a
Length=1498
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 61/162 (37%), Positives = 89/162 (54%), Gaps = 1/162 (0%)
Query 74 HHDHVT-AGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQL 132
+HDH GH G T A +S YYWP ++ Y+ +C C+ KS + GLL+ L
Sbjct 1129 YHDHTLFGGHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRLHGLLQPL 1188
Query 133 LIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADR 192
I R +S+DF+T LP T+ + ILV+V+ SK A F+ +K+ A ++LL
Sbjct 1189 PIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFIATRKTLDATQLIDLLFRY 1248
Query 193 LIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ YH FP+ + SDRD R +D + +L R IK MSS+ H
Sbjct 1249 IFSYHGFPRTITSDRDVRMTADKYQELTKRLGIKSTMSSANH 1290
> YGR109w-b
Length=1547
Score = 117 bits (294), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 61/162 (37%), Positives = 89/162 (54%), Gaps = 1/162 (0%)
Query 74 HHDHVT-AGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQL 132
+HDH GH G T A +S YYWP ++ Y+ +C C+ KS + GLL+ L
Sbjct 1103 YHDHTLFGGHFGVTVTLAKISPIYYWPKLQHSIIQYIRTCVQCQLIKSHRPRLHGLLQPL 1162
Query 133 LIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADR 192
I R +S+DF+T LP T+ + ILV+V+ SK A F+ +K+ A ++LL
Sbjct 1163 PIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFIATRKTLDATQLIDLLFRY 1222
Query 193 LIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ YH FP+ + SDRD R +D + +L R IK MSS+ H
Sbjct 1223 IFSYHGFPRTITSDRDVRMTADKYQELTKRLGIKSTMSSANH 1264
> At2g10780
Length=1611
Score = 112 bits (281), Expect = 5e-25, Method: Composition-based stats.
Identities = 56/179 (31%), Positives = 90/179 (50%), Gaps = 1/179 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
R+CVP + L H + H G K + L +Y+W GM+ +V C C+
Sbjct 1150 RVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQ 1209
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPL-TTTGHDSILVMVESLSKMADFVR 175
K+ +Q P+GLL+ L IP + H+++DF+T LP + H+++ V+V+ L+K A F+
Sbjct 1210 LVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMA 1269
Query 176 AKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
A E D ++R H P ++SDRD RF S W + +S++YH
Sbjct 1270 ISDKDGAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYH 1328
> At2g07660
Length=949
Score = 112 bits (279), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 60/186 (32%), Positives = 94/186 (50%), Gaps = 3/186 (1%)
Query 50 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYV 109
I V+G R+CVP + L H + H G K + L +Y+W GMR +V
Sbjct 537 IVVNG--RVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMRKDVARWV 594
Query 110 ESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPL-TTTGHDSILVMVESLS 168
C C+ K+ +Q P+GLL+ L I + H+++DF+T LP + H+++ V+V+ L+
Sbjct 595 AKCPTCQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLT 654
Query 169 KMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRC 228
K A F+ A E D ++R H P ++SDRD RF S WN +
Sbjct 655 KSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVN 714
Query 229 MSSSYH 234
+S++YH
Sbjct 715 LSTAYH 720
> At4g07850
Length=1138
Score = 109 bits (273), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 60/178 (33%), Positives = 88/178 (49%), Gaps = 2/178 (1%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
R+C+P + H GH G KT + +H++WP M+ E CT C+
Sbjct 750 RLCIPN-SSLRELFIREAHGGGLMGHFGVSKTLKVMQDHFHWPHMKRDVERMCERCTTCK 808
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+K+ +Q P GL L IP +S+DF+ LP T TG DSI V+V+ SKMA F+
Sbjct 809 QAKAKSQ-PHGLCTPLPIPLHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFIPC 867
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
K+ A L ++R H P+ ++SDRD +F S W L + K S++ H
Sbjct 868 HKTDDAMHIANLFFREVVRLHGMPKTIVSDRDTKFLSYFWKTLWSKLGTKLLFSTTCH 925
> At4g07830
Length=611
Score = 109 bits (272), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 60/185 (32%), Positives = 95/185 (51%), Gaps = 3/185 (1%)
Query 50 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYV 109
I VHG R+CVP+ E + L H + + H G K + L +Y W GM+ +V
Sbjct 155 ILVHG--RVCVPKDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVGNWV 212
Query 110 ESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSK 169
E C C+ K +Q LL+ L IP + +++DF+ LP++ T D+I V+V+ L+K
Sbjct 213 EECDVCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTK 271
Query 170 MADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCM 229
A F+ +K+ AA + +++ H P ++SDRD +F S W K M
Sbjct 272 SAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQM 331
Query 230 SSSYH 234
S++YH
Sbjct 332 STAYH 336
> At1g35647
Length=1495
Score = 108 bits (270), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 60/178 (33%), Positives = 88/178 (49%), Gaps = 2/178 (1%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
R+C+P + H GH G KT + +H++WP M+ E C C+
Sbjct 983 RLCIPN-SSLRELFIREAHGGGLMGHFGVSKTIKVMQDHFHWPHMKRDVERICERCPTCK 1041
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+K+ +Q P GL L IPS +S+DF+ LP T TG DSI V+V+ SKMA F+
Sbjct 1042 QAKAKSQ-PHGLYTPLPIPSHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFIPC 1100
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
K+ A L ++R H P+ ++SDRD +F S W L + K S++ H
Sbjct 1101 HKTDDAIHIANLFFREVVRLHGMPKTIVSDRDTKFLSYFWKTLWSKLGTKLLFSTTCH 1158
> At1g35370_2
Length=923
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 59/178 (33%), Positives = 87/178 (48%), Gaps = 1/178 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I VP E + L H G G+ + + +YW GM A++ SC C+
Sbjct 518 KIVVPNDVEITNKLLQWLHCSGMGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQ 577
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
KS N GLL+ L IP + VS+DFI LP + G I+V+V+ LSK A FV
Sbjct 578 QCKSDNAAYPGLLQPLPIPDKIWCDVSMDFIEGLP-NSGGKSVIMVVVDRLSKAAHFVAL 636
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
++A + D + ++H P ++SDRD F SD W + ++ MSS+YH
Sbjct 637 AHPYSALTVAQAFLDNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYH 694
> At3g31970
Length=1329
Score = 102 bits (255), Expect = 5e-22, Method: Composition-based stats.
Identities = 58/185 (31%), Positives = 93/185 (50%), Gaps = 3/185 (1%)
Query 50 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYV 109
I VHG R+CVP+ E + L H + + H K + L +Y W GM+ +V
Sbjct 888 ILVHG--RVCVPKDEELRREILSEAHASMFSIHPRATKMYRDLKRYYQWVGMKRDVANWV 945
Query 110 ESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSK 169
C C+ K+ +Q P GLL+ L I + +++DF+ LP++ T D+I V+V+ L+K
Sbjct 946 TECDVCQLVKAEHQVPGGLLQSLPILEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTK 1004
Query 170 MADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCM 229
A F+ +K+ A + ++ H P ++SDRD +F S W K M
Sbjct 1005 SAHFLAIRKTDGAVLLAKKYVSEIVELHGVPVSIVSDRDSKFTSAFWRAFQGEMGTKVQM 1064
Query 230 SSSYH 234
S++YH
Sbjct 1065 STAYH 1069
> At4g16910
Length=687
Score = 97.4 bits (241), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 55/186 (29%), Positives = 87/186 (46%), Gaps = 11/186 (5%)
Query 50 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYV 109
I V+G R+C P + L H + H G K + L +Y+W GM+ +V
Sbjct 237 IVVNG--RVCGPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV 294
Query 110 ESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPL-TTTGHDSILVMVESLS 168
H Q P+G+L+ L IP + H+ +DF+T LP + H+++ V+V+ L+
Sbjct 295 AKEEH--------QVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLT 346
Query 169 KMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRC 228
K A F+ A E D ++R H P ++SDRD RF S W +
Sbjct 347 KSAHFMAISDKDAAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVN 406
Query 229 MSSSYH 234
+S++YH
Sbjct 407 LSTAYH 412
> SPAC167.08
Length=1214
Score = 96.7 bits (239), Expect = 4e-20, Method: Composition-based stats.
Identities = 54/177 (30%), Positives = 87/177 (49%), Gaps = 1/177 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I +P + + +H+ H G + + + W G+R YV++C C+
Sbjct 783 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 842
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+KS N KP G L+ + R +S+DFIT LP ++G++++ V+V+ SKMA V
Sbjct 843 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPC 901
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
KS TA T + R+I Y P+ +I+D D F S W H++N S Y
Sbjct 902 TKSITAEQTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPY 958
> SPAC9.04
Length=1333
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 54/177 (30%), Positives = 87/177 (49%), Gaps = 1/177 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I +P + + +H+ H G + + + W G+R YV++C C+
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+KS N KP G L+ + R +S+DFIT LP ++G++++ V+V+ SKMA V
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPC 1020
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
KS TA T + R+I Y P+ +I+D D F S W H++N S Y
Sbjct 1021 TKSITAEQTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPY 1077
> SPAC26A3.13c
Length=1333
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 54/177 (30%), Positives = 87/177 (49%), Gaps = 1/177 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I +P + + +H+ H G + + + W G+R YV++C C+
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+KS N KP G L+ + R +S+DFIT LP ++G++++ V+V+ SKMA V
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPC 1020
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
KS TA T + R+I Y P+ +I+D D F S W H++N S Y
Sbjct 1021 TKSITAEQTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPY 1077
> SPAC13D1.01c
Length=1333
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 54/177 (30%), Positives = 87/177 (49%), Gaps = 1/177 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I +P + + +H+ H G + + + W G+R YV++C C+
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+KS N KP G L+ + R +S+DFIT LP ++G++++ V+V+ SKMA V
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPC 1020
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
KS TA T + R+I Y P+ +I+D D F S W H++N S Y
Sbjct 1021 TKSITAEQTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPY 1077
> SPCC1020.14
Length=1333
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 54/177 (30%), Positives = 87/177 (49%), Gaps = 1/177 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I +P + + +H+ H G + + + W G+R YV++C C+
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+KS N KP G L+ + R +S+DFIT LP ++G++++ V+V+ SKMA V
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPC 1020
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
KS TA T + R+I Y P+ +I+D D F S W H++N S Y
Sbjct 1021 TKSITAEQTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPY 1077
> SPBC1289.17
Length=1333
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 54/177 (30%), Positives = 87/177 (49%), Gaps = 1/177 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I +P + + +H+ H G + + + W G+R YV++C C+
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+KS N KP G L+ + R +S+DFIT LP ++G++++ V+V+ SKMA V
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPC 1020
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
KS TA T + R+I Y P+ +I+D D F S W H++N S Y
Sbjct 1021 TKSITAEQTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPY 1077
> SPAC2E1P3.03c
Length=1333
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 54/177 (30%), Positives = 87/177 (49%), Gaps = 1/177 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I +P + + +H+ H G + + + W G+R YV++C C+
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+KS N KP G L+ + R +S+DFIT LP ++G++++ V+V+ SKMA V
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPC 1020
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
KS TA T + R+I Y P+ +I+D D F S W H++N S Y
Sbjct 1021 TKSITAEQTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPY 1077
> SPAC27E2.08
Length=1333
Score = 96.3 bits (238), Expect = 5e-20, Method: Composition-based stats.
Identities = 54/177 (30%), Positives = 87/177 (49%), Gaps = 1/177 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I +P + + +H+ H G + + + W G+R YV++C C+
Sbjct 902 QILLPNDTQLTRTIIKKYHEEGKLIHPGIELLTNIILRRFTWKGIRKQIQEYVQNCHTCQ 961
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
+KS N KP G L+ + R +S+DFIT LP ++G++++ V+V+ SKMA V
Sbjct 962 INKSRNHKPYGPLQPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPC 1020
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
KS TA T + R+I Y P+ +I+D D F S W H++N S Y
Sbjct 1021 TKSITAEQTARMFDQRVIAYFGNPKEIIADNDHIFTSQTWKDFAHKYNFVMKFSLPY 1077
> At3g11970_2
Length=958
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 71/139 (51%), Gaps = 1/139 (0%)
Query 96 YYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTT 155
+YW GM AY+ SC C+ KS GLL+ L IP + VS+DFI LP+ +
Sbjct 569 FYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPV-SG 627
Query 156 GHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDL 215
G I+V+V+ LSK A F+ ++A D + + H P ++SDRD F S+
Sbjct 628 GKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGCPTSIVSDRDVVFTSEF 687
Query 216 WNQLCHRFNIKRCMSSSYH 234
W + + ++S+YH
Sbjct 688 WREFFTLQGVALKLTSAYH 706
> At1g36590_2
Length=958
Score = 83.2 bits (204), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 46/139 (33%), Positives = 71/139 (51%), Gaps = 1/139 (0%)
Query 96 YYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTT 155
+Y GM AY+ SC C+ KS GLL+ L IP + VS+DFI LP+ +
Sbjct 569 FYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPV-SG 627
Query 156 GHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDL 215
G I+V+V+ LSK A F+ ++A + D + + H P ++SDRD F S+
Sbjct 628 GKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFKLHGCPTSIVSDRDVVFTSEF 687
Query 216 WNQLCHRFNIKRCMSSSYH 234
W + + ++S+YH
Sbjct 688 WREFFTLQGVALKLTSAYH 706
> At2g14650
Length=1328
Score = 83.2 bits (204), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 41/127 (32%), Positives = 70/127 (55%), Gaps = 1/127 (0%)
Query 108 YVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESL 167
+V C C+ K+ +Q P G+L+ L IP + +++DF+ LP++ T D+I V+V+ L
Sbjct 946 WVAECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRT-KDAIWVIVDRL 1004
Query 168 SKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKR 227
+K A F+ +K+ AA + +++ H P ++SDRD +F S W K
Sbjct 1005 TKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKV 1064
Query 228 CMSSSYH 234
MS++YH
Sbjct 1065 QMSTAYH 1071
> At2g05610
Length=780
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 51/160 (31%), Positives = 75/160 (46%), Gaps = 2/160 (1%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+I VP L H GH G++ T + +YW M A++ SC C+
Sbjct 462 KIVVPNNSGIKDTILRWLHCSGMGGHSGKEVTHQRVKGLFYWKSMVKDIQAFIRSCGTCQ 521
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
KS N GLL+ L IP R + VS+DFI LPL + G I+V+V+ LSK A F+
Sbjct 522 QCKSDNAASPGLLQPLPIPDRIWSDVSMDFIDGLPL-SNGKTVIMVVVDRLSKAAHFIAL 580
Query 177 KKSFTAADTVELLADRLIRYHSFP-QVLISDRDPRFQSDL 215
++A + D + + H P +++S R Q L
Sbjct 581 AHPYSAMTVAQAYLDNVFKLHGCPSSIVVSSRRVLVQHQL 620
> At2g04670
Length=1411
Score = 73.6 bits (179), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 47/185 (25%), Positives = 78/185 (42%), Gaps = 34/185 (18%)
Query 50 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYV 109
I V+G R+CVP+ E + L H + + H G K + L +Y W GM+ +V
Sbjct 1001 IFVYG--RVCVPKDEELRREILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVANWV 1058
Query 110 ESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSK 169
C C+ K+ +Q P D+I V+++ L+K
Sbjct 1059 AECDVCQLVKAEHQVP--------------------------------DAIWVIMDRLTK 1086
Query 170 MADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCM 229
A F+ +K+ AA + +++ H P ++SDRD +F W + K M
Sbjct 1087 SAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQM 1146
Query 230 SSSYH 234
S++YH
Sbjct 1147 STAYH 1151
> At2g06190
Length=280
Score = 67.0 bits (162), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 50/91 (54%), Gaps = 0/91 (0%)
Query 144 LDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVL 203
+DF+ LP T G DS+ V+V+ SKM F+ KK+ A++ +L ++R H P+ +
Sbjct 1 MDFVLGLPRTQRGVDSVFVVVDRFSKMTHFIACKKTADASNIAKLFFKEVVRLHGVPKSI 60
Query 204 ISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
SDRD +F S W+ L F SS+ H
Sbjct 61 TSDRDTKFLSHFWSTLWRMFGTALNRSSTPH 91
> At4g03840
Length=973
Score = 64.3 bits (155), Expect = 2e-10, Method: Composition-based stats.
Identities = 29/90 (32%), Positives = 46/90 (51%), Gaps = 0/90 (0%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
R+CVP + L H + H G K + L +Y+W GM+ +V C C+
Sbjct 600 RVCVPNNRALKEEILREAHQSKFSIHPGSNKIYRDLKRYYHWVGMKKDVARWVAKCPTCQ 659
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDF 146
K+ +Q P+GLL+ L IP + H+++DF
Sbjct 660 LVKAEHQVPSGLLQNLPIPEWKWDHITMDF 689
> At1g36120
Length=1235
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 51/208 (24%), Positives = 77/208 (37%), Gaps = 48/208 (23%)
Query 27 REDLQIEFCNRQFTFRYLQPYLHIRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQK 86
RE L++E +R L VH R+C+P+ E + L H + + H G
Sbjct 845 REPLKLEAVDRAANGTIL-------VHE--RVCLPKDEELRREILSEAHASMFSIHPGAT 895
Query 87 KTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDF 146
K + L HY W GM+ +V C C+ K+ +Q P GLL+ L I
Sbjct 896 KMYRDLKRHYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSLPISE---------- 945
Query 147 ITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISD 206
KK+ AA + +++ H P ++S
Sbjct 946 -----------------------------WKKTDGAAVLPKKYVSEIVKLHGVPVSILSH 976
Query 207 RDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
RD +F S W K MS++YH
Sbjct 977 RDSKFTSAFWRAFQVEMGTKVQMSTAYH 1004
> At2g06470
Length=899
Score = 63.9 bits (154), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 52/105 (49%), Gaps = 2/105 (1%)
Query 50 IRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYV 109
I V+G R+CVP + L H + H G K + L +Y+W GM+ +V
Sbjct 595 IVVNG--RVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV 652
Query 110 ESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTT 154
C C+ K+ +Q P+GLL+ L IP + H+++DF L T
Sbjct 653 AKCPTCQLVKAEHQVPSGLLQNLPIPEWKWDHITMDFAFQKALGT 697
> At4g10580
Length=1240
Score = 59.7 bits (143), Expect = 5e-09, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 59/114 (51%), Gaps = 1/114 (0%)
Query 87 KTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLDF 146
K + L +Y W GM+ +V C C+ K+ +Q G+L+ L IP + +++D
Sbjct 960 KMYRDLKRYYQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDL 1019
Query 147 ITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFP 200
+ L ++ T D+I V+V+ L+K A F+ +K+ AA + +++ H P
Sbjct 1020 VVGLRVSRT-KDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVP 1072
> At2g06890
Length=1215
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 37/107 (34%), Positives = 49/107 (45%), Gaps = 4/107 (3%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
R+CVP + H GH G KT ++EH+ WP M+ C C+
Sbjct 884 RLCVPNC-SLRDLFVREAHGGGLMGHFGIAKTLEVMTEHFRWPHMKCDVKRICGRCNTCK 942
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVM 163
+KS Q P GL L IP +S+DF+ LP TG DSI V+
Sbjct 943 QAKSKIQ-PNGLYTPLPIPKHPWNDISMDFVMGLP--RTGKDSIFVV 986
> CE14062_2
Length=812
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 44/178 (24%), Positives = 82/178 (46%), Gaps = 15/178 (8%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
R+ VP+ + L H+ GH G + +W G+ V C +C+
Sbjct 555 RVVVPK--SLQMKVLRQLHE----GHPGVVRMKQKACSFVFWTGIDKDVEKLVRGCENCQ 608
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
S + + L P + + V +DF + GH LV+V++ SK A+ V+
Sbjct 609 ESAKMPR--VAPLRPWPEPQKAWSRVHIDFAGPV----NGH-WFLVIVDAKSKYAE-VKM 660
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
K+ +A+ TV LL + + H +P++L+SD +F S+ + +C + ++ S+ Y+
Sbjct 661 TKTISASATVSLL-EEVFATHGYPELLVSDNGTQFTSNQFKLMCQEYGMEHKTSAVYY 717
> At2g13020
Length=930
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 51/230 (22%), Positives = 105/230 (45%), Gaps = 24/230 (10%)
Query 17 VPYKAAANQPREDLQIEFCNRQFT-----FRYLQPYLHIRVH-GLWRICVP--QFPEFLT 68
V Y AA +P D ++ ++F +++ +PYL+ + G++R C+ + P L+
Sbjct 143 VNYLAADVEP--DNFTDYNKKRFLREIRRYQWDEPYLYKHSYDGIYRRCIAATEVPSILS 200
Query 69 QTLHSHHDHVTAGHQGQKKTFA-ALSEHYYWPGMRAYTTAYVESCTHCRASKSL---NQK 124
H GH KT + L ++WP M T ++ C C+ + N+
Sbjct 201 HC----HSSSYGGHFATFKTVSKVLQADFWWPTMFRDTQKFISQCDPCQRRGKISKRNEM 256
Query 125 PAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAAD 184
P ++ + R +DF+ P + + ILV V+ +SK + + + K+ +A
Sbjct 257 PPNFRLEVEVFDR----WGIDFMGPFP-PSNKNLYILVDVDYVSKWVEAIASLKNDSAV- 310
Query 185 TVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
++L + P+++ISD D F + + +L ++ ++ +++ YH
Sbjct 311 VMKLFKSIIFPRFGVPRIVISDGDKHFINKILEKLLLQYGVQHRVATPYH 360
> 7292616
Length=1062
Score = 51.6 bits (122), Expect = 2e-06, Method: Composition-based stats.
Identities = 31/157 (19%), Positives = 73/157 (46%), Gaps = 12/157 (7%)
Query 77 HVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPS 136
H+ H+G +T + + Y+P ++ + + C C+ K Q + + P+
Sbjct 738 HIKNNHRGIDETVSHIKRQIYFPCLKERVSQLINKCDICQTLKYDRQPQKPIFQLTETPN 797
Query 137 RRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLI-- 194
+ LD I + L + + +IL +++ SK A+ + + D++ + +
Sbjct 798 K-----PLD-IVHIDLYSINNKTILTIIDKFSKFAE----GYTIPSRDSINITKHMMFFF 847
Query 195 RYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSS 231
+ H P+ ++ D+ P F ++ +LC+++NI ++S
Sbjct 848 KTHGIPKTIVCDQGPEFAGIIFKELCNQYNITLHVTS 884
> CE02792_2
Length=634
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 42/179 (23%), Positives = 84/179 (46%), Gaps = 17/179 (9%)
Query 57 RICVPQ-FPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHC 115
R+ VP+ + + + LH H + Q + ++F +W G+ + V C +C
Sbjct 376 RVIVPKSLQKIVLKQLHEGHPGIVQMKQ-KARSFV------FWRGLDSDIENMVRHCNNC 428
Query 116 RASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVR 175
+ + + + L +P + +DF PL +LV+V++ +K A+ V+
Sbjct 429 QENSKMPRVVP--LNPWPVPEAPWKRIHIDFAG--PLNGC---YLLVVVDAKTKYAE-VK 480
Query 176 AKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+S +A T++LL + + H +P+ +ISD + S L+ Q+C I+ S+ Y+
Sbjct 481 LTRSISAVTTIDLL-EEIFSIHGYPETIISDNGTQLTSHLFAQMCQSHGIEHKTSAVYY 538
> CE29321
Length=2186
Score = 49.7 bits (117), Expect = 6e-06, Method: Composition-based stats.
Identities = 45/182 (24%), Positives = 76/182 (41%), Gaps = 13/182 (7%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
R VP+ + T L H+ + AGH G KK + + +YWP MR V +C C
Sbjct 1457 RSVVPE--KIRTPLLKELHEGMLAGHFGIKKMWRMVHRKFYWPQMRVCVENCVRTCAKCL 1514
Query 117 ASKSLNQKPAGLLEQLLIPSRRR---AHVSLDFITDLPLTTTGHDSILVMVESLSKMADF 173
+ ++ L L P R V+ D + D+ L+ G+ IL +++ +K
Sbjct 1515 CANDHSK-----LTSSLTPYRMTFPLEIVACDLM-DVGLSVQGNRYILTIIDLFTKYGTA 1568
Query 174 VRAKKSFTAADTVELLADR-LIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSS 232
V A ++ +R I P L++D+ F + L+ Q H I+ +
Sbjct 1569 VPIPDK-KAETVLKAFVERWAIGEGRIPLKLLTDQGKEFVNGLFAQFTHMLKIEHITTKG 1627
Query 233 YH 234
Y+
Sbjct 1628 YN 1629
> CE07254
Length=2175
Score = 49.7 bits (117), Expect = 6e-06, Method: Composition-based stats.
Identities = 45/182 (24%), Positives = 76/182 (41%), Gaps = 13/182 (7%)
Query 57 RICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCR 116
R VP+ + T L H+ + AGH G KK + + +YWP MR V +C C
Sbjct 1446 RSVVPE--KIRTPLLKELHEGMLAGHFGIKKMWRMVHRKFYWPQMRVCVENCVRTCAKCL 1503
Query 117 ASKSLNQKPAGLLEQLLIPSRRR---AHVSLDFITDLPLTTTGHDSILVMVESLSKMADF 173
+ ++ L L P R V+ D + D+ L+ G+ IL +++ +K
Sbjct 1504 CANDHSK-----LTSSLTPYRMTFPLEIVACDLM-DVGLSVQGNRYILTIIDLFTKYGTA 1557
Query 174 VRAKKSFTAADTVELLADR-LIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSS 232
V A ++ +R I P L++D+ F + L+ Q H I+ +
Sbjct 1558 VPIPDK-KAETVLKAFVERWAIGEGRIPLKLLTDQGKEFVNGLFAQFTHMLKIEHITTKG 1616
Query 233 YH 234
Y+
Sbjct 1617 YN 1618
> CE13102_2
Length=813
Score = 49.3 bits (116), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 37/154 (24%), Positives = 72/154 (46%), Gaps = 9/154 (5%)
Query 81 GHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRA 140
GH G + +W G+ + V C +C+ + + + L +P
Sbjct 573 GHPGIVQMKQKARAFVFWRGLDSDIEKMVRHCNNCQENSKMPRVVP--LNPWPVPETPWK 630
Query 141 HVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFP 200
+ +DF L G+ +LV+V++ +K A+ V+ +S +A T++LL + + H +P
Sbjct 631 RIHIDFAGPL----NGY-YLLVVVDAKTKYAE-VKLTRSISAVTTIDLL-EEIFSIHGYP 683
Query 201 QVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ +ISD + S L+ Q+C I+ S+ Y+
Sbjct 684 ETIISDNGTQLTSHLFAQMCQSHGIEHKTSAVYY 717
> Hs22049976
Length=477
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 48/182 (26%), Positives = 76/182 (41%), Gaps = 17/182 (9%)
Query 57 RICVPQF--PEFLTQTLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTH 114
R+ +P+ P+F+ Q H H G+ + H+Y P + A T A E C
Sbjct 173 RLAIPEAIAPQFMKQ-FHQ------GTHMGKTALETLVGWHFYVPCLTAITRAVCEQCLT 225
Query 115 CRASK--SLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMAD 172
C + + +P G+ E P ++ +DF T+LP G+ +LV V + S +
Sbjct 226 CAQNNPWQVPTQPPGIQETGATPCE---NLLVDF-TELP-RARGYQYMLVFVCTFSGWVE 280
Query 173 FVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSS 232
+ T LL D + R+ P L SD P F +++ QL IK +
Sbjct 281 AFPTRIEKAQEVTRLLLKDIIPRF-GLPLTLGSDNGPAFMAEVVQQLSQLLKIKWKLHIV 339
Query 233 YH 234
YH
Sbjct 340 YH 341
> At2g06320
Length=466
Score = 45.8 bits (107), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 38/164 (23%), Positives = 72/164 (43%), Gaps = 10/164 (6%)
Query 75 HDHVTAGHQGQKKTFAA-LSEHYYWPGMRAYTTAYVESCTHCRASKSL---NQKPAGLLE 130
HD GH KT + L ++WP M +V C C+ ++ N+ P +
Sbjct 104 HDSAYGGHFATFKTVSKILQAGFWWPTMFKDAQEFVSKCDSCQRKGNISRRNEMPQNPIL 163
Query 131 QLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLA 190
++ I +DF+ P ++ G+ ILV+V+ +SK + + A + A ++L
Sbjct 164 EVDIFDVW----GIDFMGPFP-SSYGNKYILVVVDYVSKWVEAI-ASPTNDAKVVLKLFK 217
Query 191 DRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+ V+ISD F + ++ L + +K +++ YH
Sbjct 218 TIIFPRFGVSWVVISDGGKHFINKVFENLLKKHGVKHKVATPYH 261
> At1g42375
Length=1773
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 46/195 (23%), Positives = 86/195 (44%), Gaps = 13/195 (6%)
Query 45 QPYLHIRVHG-LWRICVPQFPEFLTQTLHSHHDHVTAGHQGQKKTFAA-LSEHYYWPGMR 102
+PYL+ ++R CV + E LH H GH KT + L ++WP M
Sbjct 1382 EPYLYTLCKDKIYRRCVSE-DEVEGILLHCHG-SAYGGHFATFKTVSKILQAGFWWPTMF 1439
Query 103 AYTTAYVESCTHCRASKSL---NQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDS 159
+V C C+ ++ N+ P + ++ I +DF+ P ++ G+
Sbjct 1440 KDAQEFVSKCDSCQRKGNISRRNEMPQNPILEVEIFDVW----GIDFMGPFP-SSYGNKY 1494
Query 160 ILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQL 219
ILV V+ +SK + + A + A ++L + P+V+ISD F + ++ L
Sbjct 1495 ILVAVDYVSKWVEAI-ASPTNDAKVVLKLFKTIIFPRFGVPRVVISDGGKHFINKVFENL 1553
Query 220 CHRFNIKRCMSSSYH 234
+ +K +++ Y+
Sbjct 1554 LKKHGVKHKVATPYN 1568
> At2g15100
Length=1329
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 37/170 (21%), Positives = 63/170 (37%), Gaps = 19/170 (11%)
Query 58 ICVPQFPEFLTQTLHSHHDHVTAGHQGQKK-TFAALSEHYYWPGMRAYTTAYVESCTHCR 116
+C+ F E + HD H G + F YYWP + A AY C C+
Sbjct 979 VCI--FGEQTRTVMKEVHDGTCGNHTGGRSLAFKVRKYGYYWPTLVADCEAYARKCEQCQ 1036
Query 117 ASKSLNQKPAGLLEQLLIPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRA 176
L +PA LL + P + +D + L ++T G ++ A
Sbjct 1037 KHAPLILQPAELLTTVSAPYPFMKWL-MDIVGPLHVSTRGVEAA---------------A 1080
Query 177 KKSFTAADTVELLADRLIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIK 226
+ T + +I H P +++D +F S+ + C + I+
Sbjct 1081 YSNITHVQVWNFIWKDIICRHGLPYEIVTDNGSQFISEQFEVFCEEWQIR 1130
> Hs8923124
Length=392
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 40/206 (19%), Positives = 83/206 (40%), Gaps = 5/206 (2%)
Query 26 PREDLQIEFCNRQFTFRYLQPYLHIRVHGLWRICVPQFPEFLTQTLHSHHDHVTAGHQGQ 85
P E I ++F F+ + + + R+ + E + L H++ + H G
Sbjct 30 PSERSGIRRAAKKFVFKEKKLFYVGKDRKQNRLVIVSEEE-KKKVLRECHENDSGAHHGI 88
Query 86 KKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRAHVSLD 145
+T + +YYW + +V +C HC+ +K N + LL + V++D
Sbjct 89 SRTLTLVESNYYWTSVTNDAKQWVYACQHCQVAK--NTVIVAPKQHLLKVENPWSLVTVD 146
Query 146 FITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFPQVLIS 205
+ + H ++M + +K + +A++ + + + Y PQ +I
Sbjct 147 LMGPFHTSNRSHVYAIIMTDLFTKWI-VILPLCDVSASEVSKAIINIFFLYGP-PQKIIM 204
Query 206 DRDPRFQSDLWNQLCHRFNIKRCMSS 231
D+ F + +L F IK+ + S
Sbjct 205 DQRDEFIQQINIELYRLFGIKQIVIS 230
> Hs22048021_2
Length=835
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 30/144 (20%), Positives = 63/144 (43%), Gaps = 6/144 (4%)
Query 71 LHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLE 130
+ S HD HQ ++T+ L +WPGM+ + Y SC C ++L ++E
Sbjct 482 IFSVHDIPLGAHQRPEETYKKLRLLGWWPGMQEHVKDYCRSCLFC-IPRNLIGSELKVIE 540
Query 131 QLLIPSRRRA---HVSLDFITDLPLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVE 187
P R A ++ ++ + + ++ GH +L++ + ++ + K +T +
Sbjct 541 SPW-PLRSTAPWSNLQIEVVGPVTISEEGHKHVLIVADPNTRWVE-AFPLKPYTHTAVAQ 598
Query 188 LLADRLIRYHSFPQVLISDRDPRF 211
+L + P L + + P+F
Sbjct 599 VLLQHVFARWGVPVRLEAAQGPQF 622
> Hs13378147
Length=150
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 0/45 (0%)
Query 71 LHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHC 115
+ S HD HQ ++T+ L +WPGM+ + Y SC C
Sbjct 74 IFSVHDIPLGAHQRPEETYKKLRLLGWWPGMQEHVKDYCRSCLFC 118
> At2g14640_2
Length=492
Score = 34.7 bits (78), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 193 LIRYHSFPQVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSYH 234
+++ H P+ ++SD DP F S W + K MS++YH
Sbjct 189 VVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYH 230
> CE01358
Length=558
Score = 34.7 bits (78), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 22/48 (45%), Gaps = 0/48 (0%)
Query 77 HVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQK 124
H GH GQK+T + YW +R ++ SC C A K +K
Sbjct 122 HELIGHLGQKRTQMVVLRKLYWRSVRQDVKTFIASCDFCTAKKIQGRK 169
> Hs22063675
Length=163
Score = 34.7 bits (78), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 38/75 (50%), Gaps = 7/75 (9%)
Query 82 HQGQKKTFAALSEHY-YWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLLIPSRRRA 140
H GQKKT+ A++E Y + P R T ++ SCT C+ N GL + P
Sbjct 56 HAGQKKTYRAIAETYAFLP--REAVTRFLMSCTECQKRMHFNSN--GLEPKENEPPSPLV 111
Query 141 HVSLDFITDLPLTTT 155
+D+ ++PLT+T
Sbjct 112 SGIIDY--NMPLTST 124
> At2g14400
Length=1466
Score = 34.3 bits (77), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 39/96 (40%), Gaps = 2/96 (2%)
Query 75 HDHVTAGHQ-GQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSLNQKPAGLLEQLL 133
H+ + H G+ F YYWP M Y + C C+ L +P+ L +
Sbjct 979 HEGLCGSHSSGRAMAFKIKKMCYYWPTMITDCVKYAQRCKRCQLHAPLIHQPSELFSSIS 1038
Query 134 IPSRRRAHVSLDFITDLPLTTTGHDSILVMVESLSK 169
P S+D I L +T G +LV+ + SK
Sbjct 1039 APY-PFMRWSMDIIGPLHRSTRGVQYLLVLTDYFSK 1073
> Hs4505421
Length=524
Score = 32.7 bits (73), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 5/47 (10%)
Query 82 HQGQKKTFAALSEHY-YWPGMRAYTTAYVESCTHCRASKSLNQKPAG 127
H GQK+T+ A+SE Y + P R T ++ SC+ C+ LN P G
Sbjct 15 HAGQKRTYKAISESYAFLP--REAVTRFLMSCSECQKRMHLN--PDG 57
> At2g10610
Length=767
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 22/94 (23%), Positives = 38/94 (40%), Gaps = 21/94 (22%)
Query 151 PLTTTGHDSILVMVESLSKMADFVRAKKSFTAADTVELLADRLIRYHSFP---------- 200
P T+ G+ I V+++ S+ V K+ D ++ +F
Sbjct 274 PKTSVGNRYIFVLIDDYSRYMWTVLLKEK----------GDAFFKFKNFKALVEKESGEK 323
Query 201 -QVLISDRDPRFQSDLWNQLCHRFNIKRCMSSSY 233
Q I+DR F S +N C R I+R +++ Y
Sbjct 324 IQTFITDRGGEFVSGEFNSFCERTGIRRHLTAPY 357
> CE04966
Length=2037
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 35/83 (42%), Gaps = 7/83 (8%)
Query 70 TLHSHHDHVTAGHQGQKKTFAALSEHYYWPGMRAYTTAYVESCTHCRASKSL-----NQK 124
TL HV GH + T +A ++ P ++ + V +C C+ L N K
Sbjct 1581 TLVMRETHVINGHSSELYTVSAAKTMFWIPHIKVLAKSVVSNCVDCKKVHGLPFRYPNSK 1640
Query 125 PAGLLEQLLIPSRRRAHVSLDFI 147
L E+ PS+ A LD++
Sbjct 1641 --TLPEKRTSPSKPFATAGLDYM 1661
Lambda K H
0.325 0.135 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4598061294
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40