bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0275_orf3
Length=184
Score E
Sequences producing significant alignments: (Bits) Value
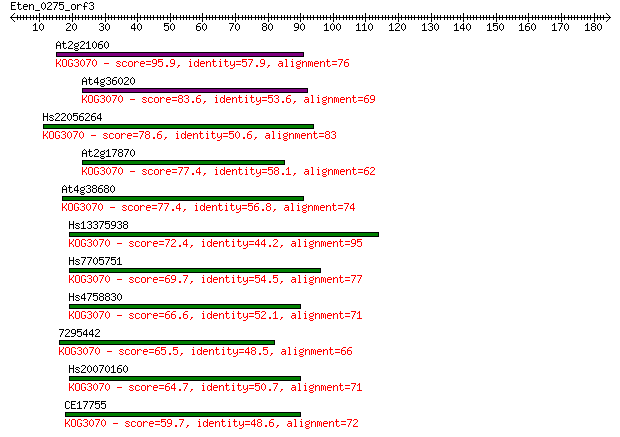
At2g21060 95.9 4e-20
At4g36020 83.6 2e-16
Hs22056264 78.6 7e-15
At2g17870 77.4 1e-14
At4g38680 77.4 1e-14
Hs13375938 72.4 5e-13
Hs7705751 69.7 3e-12
Hs4758830 66.6 3e-11
7295442 65.5 5e-11
Hs20070160 64.7 9e-11
CE17755 59.7 3e-09
> At2g21060
Length=201
Score = 95.9 bits (237), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 44/77 (57%), Positives = 59/77 (76%), Gaps = 1/77 (1%)
Query 15 DIQRGTCKWFDSKKGFGFITSDD-GTDLFVHQTEIKADGFRNLCEGEEVEFQVQSGDDGR 73
D ++GT KWFD++KGFGFIT D G DLFVHQ+ I+++GFR+L E VEF V+ + GR
Sbjct 13 DRRKGTVKWFDTQKGFGFITPSDGGDDLFVHQSSIRSEGFRSLAAEESVEFDVEVDNSGR 72
Query 74 KKAVRVTGPGGAPVKGD 90
KA+ V+GP GAPV+G+
Sbjct 73 PKAIEVSGPDGAPVQGN 89
> At4g36020
Length=299
Score = 83.6 bits (205), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 37/70 (52%), Positives = 52/70 (74%), Gaps = 1/70 (1%)
Query 23 WFDSKKGFGFITSDDGT-DLFVHQTEIKADGFRNLCEGEEVEFQVQSGDDGRKKAVRVTG 81
WF++ KG+GFIT DDG+ +LFVHQ+ I ++G+R+L G+ VEF + G DG+ KAV VT
Sbjct 17 WFNASKGYGFITPDDGSVELFVHQSSIVSEGYRSLTVGDAVEFAITQGSDGKTKAVNVTA 76
Query 82 PGGAPVKGDN 91
PGG +K +N
Sbjct 77 PGGGSLKKEN 86
> Hs22056264
Length=157
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 42/93 (45%), Positives = 58/93 (62%), Gaps = 11/93 (11%)
Query 11 EKMSDIQRGT--CKWFDSKKGFGFITS--------DDGTDLFVHQTEIKADGFRNLCEGE 60
E+ S + RGT CKWF+ + GFGFI+ D D+FVHQ+++ +GFR+L EGE
Sbjct 43 EEESQVLRGTGHCKWFNVRMGFGFISMINREGSPLDIPVDVFVHQSKLFMEGFRSLKEGE 102
Query 61 EVEFQVQSGDDGRKKAVRVTGPGGAPVKGDNRR 93
VEF + G +++RVTGPGG+P G RR
Sbjct 103 PVEFTFKKSSKGL-ESIRVTGPGGSPCLGSERR 134
> At2g17870
Length=299
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 36/63 (57%), Positives = 46/63 (73%), Gaps = 1/63 (1%)
Query 23 WFDSKKGFGFITSDDG-TDLFVHQTEIKADGFRNLCEGEEVEFQVQSGDDGRKKAVRVTG 81
WF KG+GFIT DDG +LFVHQ+ I +DGFR+L GE VE+++ G DG+ KA+ VT
Sbjct 15 WFSDGKGYGFITPDDGGEELFVHQSSIVSDGFRSLTLGESVEYEIALGSDGKTKAIEVTA 74
Query 82 PGG 84
PGG
Sbjct 75 PGG 77
> At4g38680
Length=203
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 42/75 (56%), Positives = 60/75 (80%), Gaps = 1/75 (1%)
Query 17 QRGTCKWFDSKKGFGFITSDD-GTDLFVHQTEIKADGFRNLCEGEEVEFQVQSGDDGRKK 75
++G+ KWFD++KGFGFIT DD G DLFVHQ+ I+++GFR+L E VEF+V+ ++ R K
Sbjct 11 RKGSVKWFDTQKGFGFITPDDGGDDLFVHQSSIRSEGFRSLAAEEAVEFEVEIDNNNRPK 70
Query 76 AVRVTGPGGAPVKGD 90
A+ V+GP GAPV+G+
Sbjct 71 AIDVSGPDGAPVQGN 85
> Hs13375938
Length=209
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 42/105 (40%), Positives = 58/105 (55%), Gaps = 11/105 (10%)
Query 19 GTCKWFDSKKGFGFITS--------DDGTDLFVHQTEIKADGFRNLCEGEEVEFQVQSGD 70
G CKWF+ + GFGF++ D D+FVHQ+++ +GFR+L EGE VEF +
Sbjct 42 GICKWFNVRMGFGFLSMTARAGVALDPPVDVFVHQSKLHMEGFRSLKEGEAVEFTFKKSA 101
Query 71 DGRKKAVRVTGPGGAPVKGDNRRM--SPSYNGSSRGGRGYSGGGY 113
G +++RVTGPGG G RR S+G R Y+ GG
Sbjct 102 KGL-ESIRVTGPGGVFCIGSERRPKGKSMQKRRSKGDRCYNCGGL 145
> Hs7705751
Length=364
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 42/86 (48%), Positives = 51/86 (59%), Gaps = 10/86 (11%)
Query 19 GTCKWFDSKKGFGFITSDD-GTDLFVHQTEIKADGFRNLC----EGEEVEFQVQSGDDGR 73
GT KWF+ + G+GFI +D D+FVHQT IK + R +GE VEF V G+ G
Sbjct 96 GTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKRNNPRKFLRSVGDGETVEFDVVEGEKG- 154
Query 74 KKAVRVTGPGGAPVKG----DNRRMS 95
+A VTGPGG PVKG NRR S
Sbjct 155 AEATNVTGPGGVPVKGSRYAPNRRKS 180
> Hs4758830
Length=322
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/76 (48%), Positives = 49/76 (64%), Gaps = 6/76 (7%)
Query 19 GTCKWFDSKKGFGFITSDD-GTDLFVHQTEIKADG----FRNLCEGEEVEFQVQSGDDGR 73
GT KWF+ + G+GFI +D D+FVHQT IK + R++ +GE VEF V G+ G
Sbjct 60 GTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVVEGEKG- 118
Query 74 KKAVRVTGPGGAPVKG 89
+A VTGPGG PV+G
Sbjct 119 AEAANVTGPGGVPVQG 134
> 7295442
Length=181
Score = 65.5 bits (158), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 32/67 (47%), Positives = 46/67 (68%), Gaps = 2/67 (2%)
Query 16 IQRGTCKWFDSKKGFGFITSDDG-TDLFVHQTEIKADGFRNLCEGEEVEFQVQSGDDGRK 74
++ G CKWF+ KG+GF+T +DG ++FVHQ+ I+ GFR+L E EEVEF+ Q G
Sbjct 24 VRLGKCKWFNVAKGWGFLTPNDGGQEVFVHQSVIQMSGFRSLGEQEEVEFECQRTSRGL- 82
Query 75 KAVRVTG 81
+A RV+
Sbjct 83 EATRVSS 89
> Hs20070160
Length=372
Score = 64.7 bits (156), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 36/76 (47%), Positives = 48/76 (63%), Gaps = 6/76 (7%)
Query 19 GTCKWFDSKKGFGFITSDD-GTDLFVHQTEIKADG----FRNLCEGEEVEFQVQSGDDGR 73
GT KWF+ + G+GFI +D D+FVHQT IK + R++ +GE VEF V G+ G
Sbjct 93 GTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVVEGEKG- 151
Query 74 KKAVRVTGPGGAPVKG 89
+A VTGP G PV+G
Sbjct 152 AEAANVTGPDGVPVEG 167
> CE17755
Length=208
Score = 59.7 bits (143), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 35/77 (45%), Positives = 46/77 (59%), Gaps = 6/77 (7%)
Query 18 RGTCKWFDSKKGFGFIT-SDDGTDLFVHQTEIKADG----FRNLCEGEEVEFQVQSGDDG 72
+GT KWF+ K G+GFI +D D+FVHQT I + R+L + EEV F + G G
Sbjct 22 KGTVKWFNVKNGYGFINRTDTNEDIFVHQTAIINNNPNKYLRSLGDNEEVMFDIVEGSKG 81
Query 73 RKKAVRVTGPGGAPVKG 89
+A VTGP G PV+G
Sbjct 82 L-EAASVTGPDGGPVQG 97
Lambda K H
0.311 0.145 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2950576972
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40